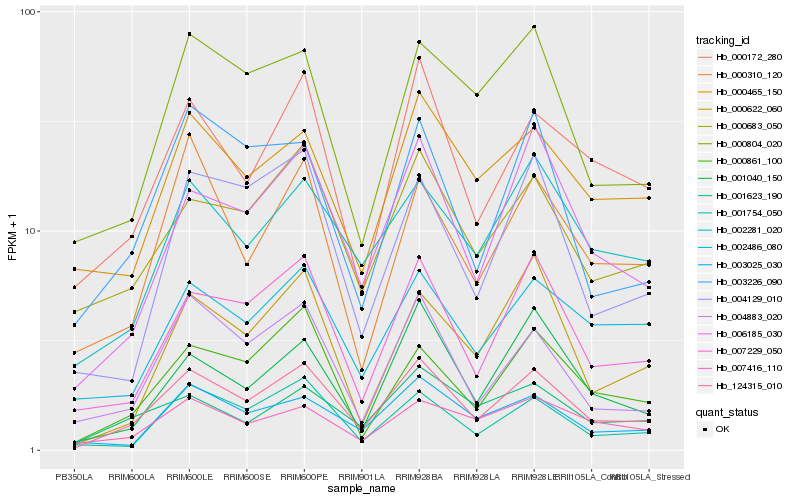
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124315_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |
| 2 |
Hb_007229_050 |
0.1080477177 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006185_030 |
0.1086560322 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 4 |
Hb_000172_280 |
0.1176987062 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 5 |
Hb_004883_020 |
0.1192080247 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 6 |
Hb_000683_050 |
0.120645474 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 7 |
Hb_000861_100 |
0.1217833895 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002281_020 |
0.1254542822 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 9 |
Hb_001754_050 |
0.1283469842 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_003025_030 |
0.1293085205 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_002486_080 |
0.1304431017 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 12 |
Hb_007416_110 |
0.1333608938 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001040_150 |
0.1350865343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001623_190 |
0.1352930678 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 15 |
Hb_000465_150 |
0.1353313647 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 16 |
Hb_003226_090 |
0.1358435188 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 17 |
Hb_000310_120 |
0.1367395791 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 18 |
Hb_000804_020 |
0.1369659103 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |
| 19 |
Hb_004129_010 |
0.1370209667 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 20 |
Hb_000622_060 |
0.1372950206 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |