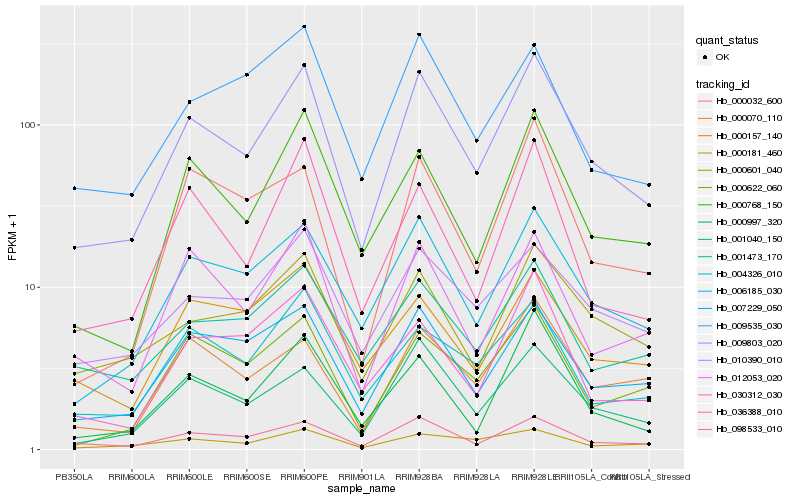
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009803_020 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 2 |
Hb_098533_010 |
0.0874201116 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 3 |
Hb_006185_030 |
0.0910427631 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 4 |
Hb_001040_150 |
0.0999324854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007229_050 |
0.1055976403 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000768_150 |
0.1127000827 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 7 |
Hb_000601_040 |
0.1177353079 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 8 |
Hb_010390_010 |
0.1178746662 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 9 |
Hb_012053_020 |
0.1209134534 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004326_010 |
0.1240835728 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_001473_170 |
0.1271047183 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 12 |
Hb_000181_460 |
0.1271600662 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000070_110 |
0.1271768759 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000997_320 |
0.1276636291 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 15 |
Hb_036388_010 |
0.1296788114 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 16 |
Hb_000622_060 |
0.1317694304 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_009535_030 |
0.1334142224 |
- |
- |
CP2 [Hevea brasiliensis] |
| 18 |
Hb_000157_140 |
0.1362935564 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_030312_030 |
0.1379353715 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000032_600 |
0.1388118586 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |