| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
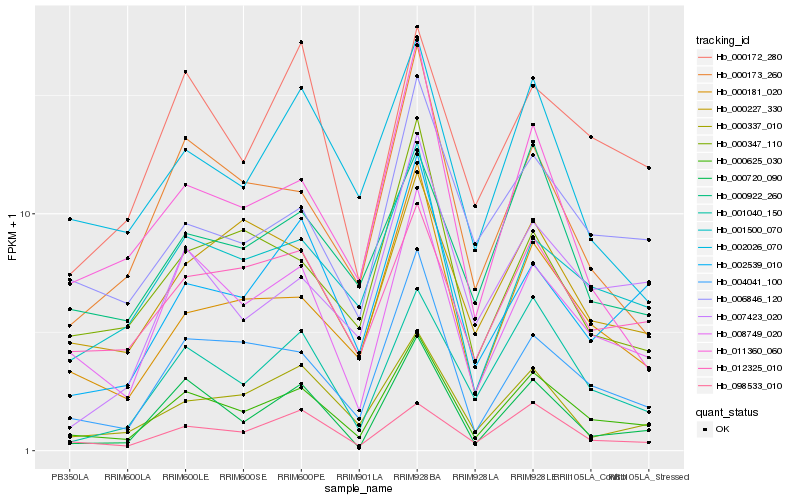
Hb_000625_030 |
0.0 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 2 |
Hb_008749_020 |
0.083977629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004041_100 |
0.1032455782 |
- |
- |
- |
| 4 |
Hb_000181_020 |
0.1111544713 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_001500_070 |
0.1211820137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001040_150 |
0.1252180543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000720_090 |
0.1272698492 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 8 |
Hb_098533_010 |
0.1279441256 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 9 |
Hb_000337_010 |
0.129661122 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_012325_010 |
0.1321619937 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 11 |
Hb_000173_260 |
0.1338304725 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 12 |
Hb_006846_120 |
0.1371923538 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 13 |
Hb_002539_010 |
0.1380430727 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 14 |
Hb_011360_060 |
0.1457531506 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 15 |
Hb_000227_330 |
0.1463755887 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 16 |
Hb_000172_280 |
0.1474281888 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 17 |
Hb_002026_070 |
0.1476293759 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 18 |
Hb_007423_020 |
0.1494805289 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 19 |
Hb_000347_110 |
0.1502254666 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 20 |
Hb_000922_260 |
0.152380102 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |