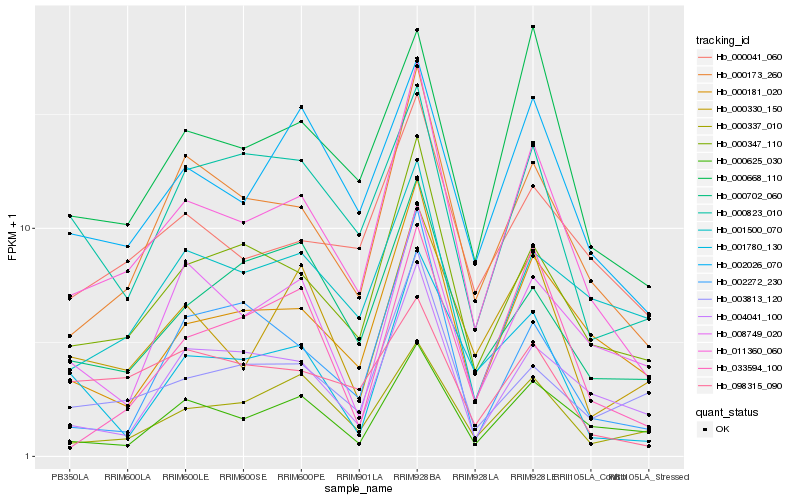
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011360_060 |
0.0 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 2 |
Hb_000173_260 |
0.1035586403 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 3 |
Hb_000347_110 |
0.1086355848 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 4 |
Hb_000181_020 |
0.1215841099 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_002026_070 |
0.1431548874 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 6 |
Hb_000041_060 |
0.1437206346 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_000625_030 |
0.1457531506 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 8 |
Hb_004041_100 |
0.1486406766 |
- |
- |
- |
| 9 |
Hb_000668_110 |
0.149872668 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 10 |
Hb_003813_120 |
0.1582503005 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000330_150 |
0.1627730033 |
- |
- |
PREDICTED: dihydroflavonol-4-reductase-like [Jatropha curcas] |
| 12 |
Hb_000337_010 |
0.1637444145 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000702_060 |
0.1684175563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001500_070 |
0.1708323815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002272_230 |
0.1709794661 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 16 |
Hb_001780_130 |
0.1738063303 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 17 |
Hb_000823_010 |
0.1754522932 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 18 |
Hb_033594_100 |
0.1770689493 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_008749_020 |
0.1784746375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_098315_090 |
0.1792957925 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |