| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
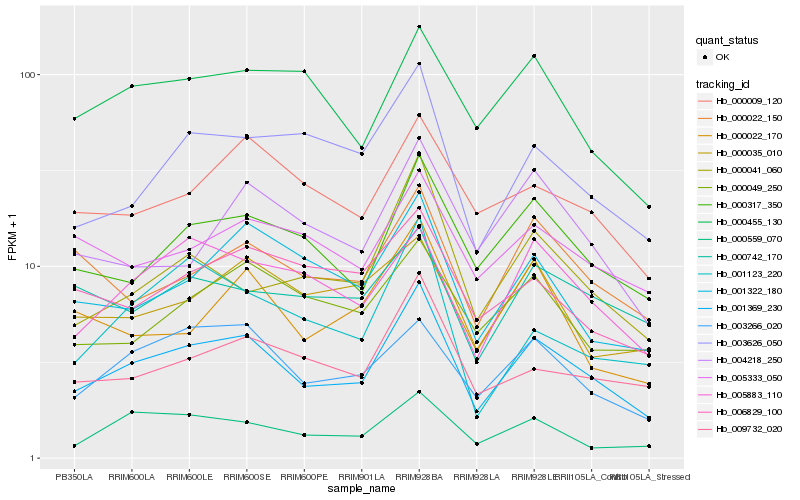
Hb_001369_230 |
0.0 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 2 |
Hb_000317_350 |
0.1178679141 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 3 |
Hb_003266_020 |
0.1264881378 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 4 |
Hb_009732_020 |
0.12698956 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 5 |
Hb_005883_110 |
0.1307556015 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000559_070 |
0.1312765776 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 7 |
Hb_000041_060 |
0.1329959467 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 8 |
Hb_003626_050 |
0.1341741869 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 9 |
Hb_001322_180 |
0.1357908643 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 10 |
Hb_000009_120 |
0.1380976768 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 11 |
Hb_000022_150 |
0.1381974319 |
- |
- |
- |
| 12 |
Hb_000022_170 |
0.1420594832 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 13 |
Hb_000049_250 |
0.1436219971 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000455_130 |
0.1445663483 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001123_220 |
0.1465591835 |
- |
- |
- |
| 16 |
Hb_005333_050 |
0.1471934398 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_004218_250 |
0.1490199393 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 18 |
Hb_000742_170 |
0.1492464859 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_006829_100 |
0.1495359387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000035_010 |
0.1500514537 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |