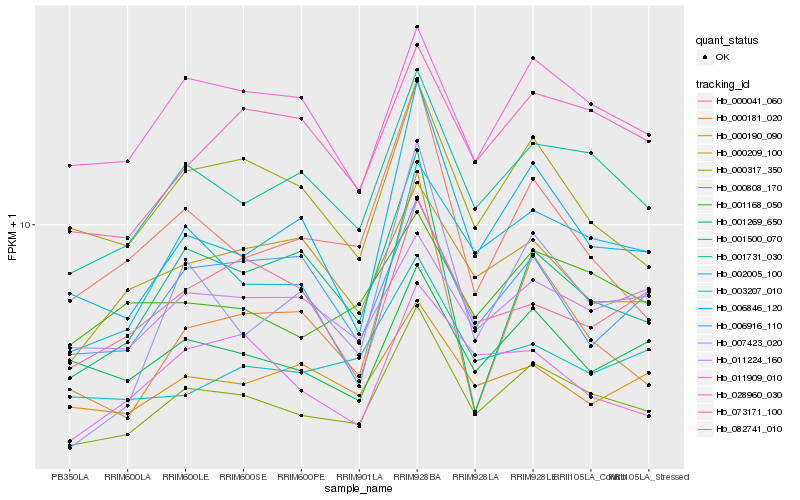
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000808_170 |
0.0 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 2 |
Hb_001731_030 |
0.0932696817 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 3 |
Hb_006846_120 |
0.1082461596 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 4 |
Hb_000041_060 |
0.1276549756 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_000317_350 |
0.1284065585 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 6 |
Hb_003207_010 |
0.1292203874 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |
| 7 |
Hb_073171_100 |
0.1358745749 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 8 |
Hb_007423_020 |
0.1380171463 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 9 |
Hb_001269_650 |
0.1392386397 |
- |
- |
transporter, putative [Ricinus communis] |
| 10 |
Hb_028960_030 |
0.141495781 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 11 |
Hb_001168_050 |
0.1451193937 |
- |
- |
hypothetical protein POPTR_0007s03190g [Populus trichocarpa] |
| 12 |
Hb_011909_010 |
0.1469706376 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 13 |
Hb_002005_100 |
0.1480554683 |
- |
- |
- |
| 14 |
Hb_000190_090 |
0.1489291407 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 15 |
Hb_001500_070 |
0.1493798387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011224_160 |
0.1534425246 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 17 |
Hb_000209_100 |
0.1545428674 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_006916_110 |
0.1552409211 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000181_020 |
0.1564454457 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_082741_010 |
0.156931461 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |