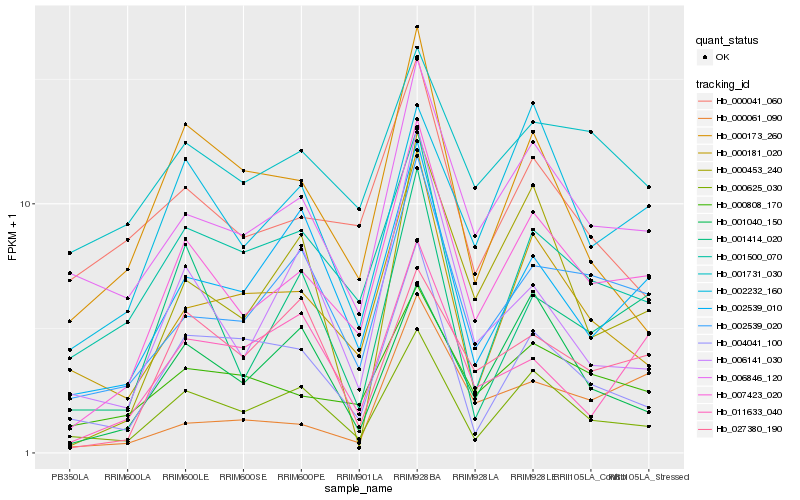
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007423_020 |
0.0 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 2 |
Hb_002539_020 |
0.1362370885 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 3 |
Hb_000808_170 |
0.1380171463 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 4 |
Hb_006846_120 |
0.1389667576 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 5 |
Hb_000625_030 |
0.1494805289 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 6 |
Hb_000181_020 |
0.1609322397 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_002539_010 |
0.1630627328 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 8 |
Hb_002232_160 |
0.1662357665 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 9 |
Hb_001414_020 |
0.1677411062 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001500_070 |
0.1697764169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004041_100 |
0.1793287906 |
- |
- |
- |
| 12 |
Hb_000041_060 |
0.1793696591 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_001731_030 |
0.1798609177 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 14 |
Hb_000453_240 |
0.1835841017 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 15 |
Hb_001040_150 |
0.1837636286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000173_260 |
0.184110742 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 17 |
Hb_006141_030 |
0.1846166624 |
- |
- |
- |
| 18 |
Hb_011633_040 |
0.1853137649 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 19 |
Hb_000061_090 |
0.1879389204 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 20 |
Hb_027380_190 |
0.188957231 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |