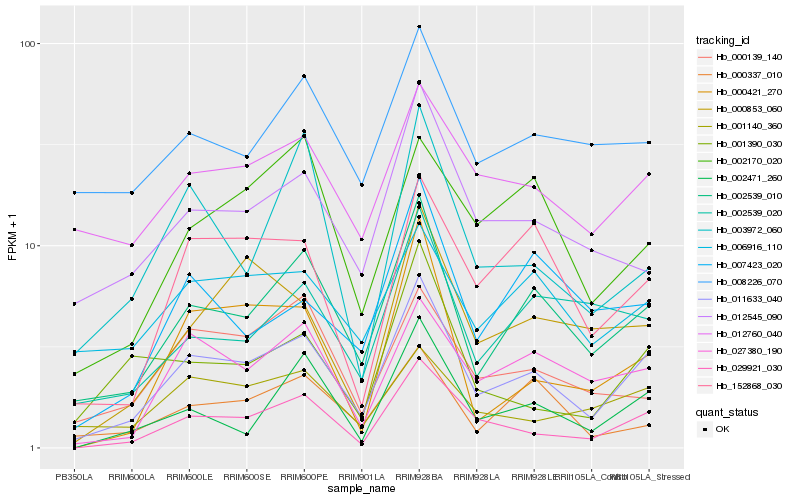
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011633_040 |
0.0 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 2 |
Hb_002539_010 |
0.1077670592 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 3 |
Hb_029921_030 |
0.1293532098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_027380_190 |
0.157672866 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 5 |
Hb_152868_030 |
0.159890709 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 6 |
Hb_012545_090 |
0.170757166 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |
| 7 |
Hb_001140_360 |
0.1707857363 |
- |
- |
- |
| 8 |
Hb_012760_040 |
0.1710786868 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 9 |
Hb_006916_110 |
0.1813417081 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002539_020 |
0.1815527549 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 11 |
Hb_000853_060 |
0.1828552362 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000139_140 |
0.1837181634 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 13 |
Hb_000337_010 |
0.1844770384 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_008226_070 |
0.1849808335 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 15 |
Hb_002170_020 |
0.1850550883 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 16 |
Hb_001390_030 |
0.1851501903 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 17 |
Hb_007423_020 |
0.1853137649 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 18 |
Hb_003972_060 |
0.1864546725 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 19 |
Hb_002471_260 |
0.1874246996 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 20 |
Hb_000421_270 |
0.1880049918 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |