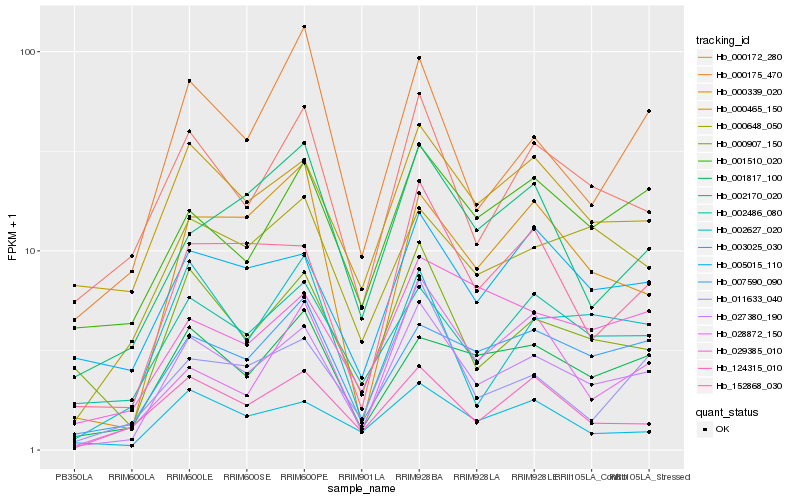
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027380_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 2 |
Hb_152868_030 |
0.1290779258 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 3 |
Hb_000172_280 |
0.1322698913 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 4 |
Hb_001817_100 |
0.1428307924 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 5 |
Hb_000465_150 |
0.1430262486 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 6 |
Hb_000907_150 |
0.1446377847 |
- |
- |
hypothetical protein L484_004295 [Morus notabilis] |
| 7 |
Hb_007590_090 |
0.1451621212 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 8 |
Hb_002486_080 |
0.1470104341 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 9 |
Hb_003025_030 |
0.1473114373 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 10 |
Hb_000648_050 |
0.1492970701 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 11 |
Hb_124315_010 |
0.150088636 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |
| 12 |
Hb_005015_110 |
0.1517177577 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 13 |
Hb_000175_470 |
0.1527664759 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 14 |
Hb_001510_020 |
0.1535473633 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 15 |
Hb_002627_020 |
0.1552568221 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 16 |
Hb_011633_040 |
0.157672866 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 17 |
Hb_000339_020 |
0.1580508449 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 18 |
Hb_002170_020 |
0.1619522039 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 19 |
Hb_029385_010 |
0.1631685012 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 20 |
Hb_028872_150 |
0.1634617112 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |