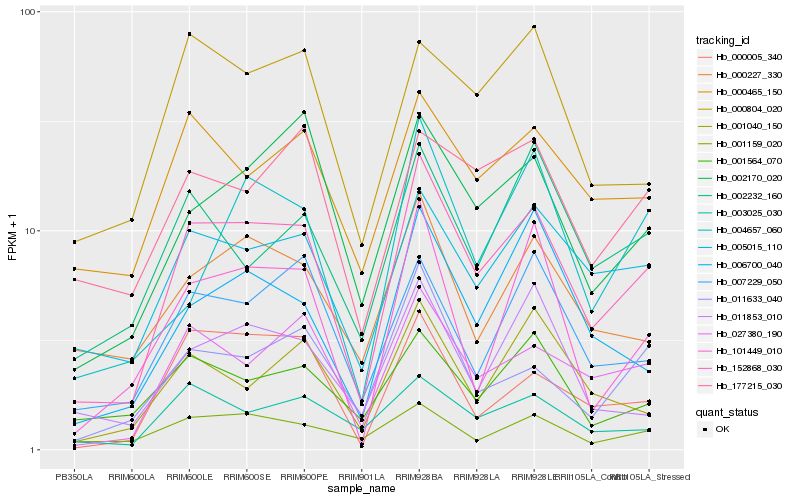
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_152868_030 |
0.0 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 2 |
Hb_027380_190 |
0.1290779258 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005015_110 |
0.1366345336 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 4 |
Hb_002170_020 |
0.1514294801 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 5 |
Hb_003025_030 |
0.1525417912 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_000465_150 |
0.1569421014 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 7 |
Hb_011633_040 |
0.159890709 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 8 |
Hb_007229_050 |
0.1607414719 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000227_330 |
0.1611200356 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 10 |
Hb_000005_340 |
0.1625643716 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006700_040 |
0.1644210523 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 12 |
Hb_101449_010 |
0.1660019017 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 13 |
Hb_002232_160 |
0.1670647279 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 14 |
Hb_001159_020 |
0.1672222564 |
- |
- |
- |
| 15 |
Hb_000804_020 |
0.1672901841 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_001564_070 |
0.1677099635 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 17 |
Hb_004657_060 |
0.1678650893 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 18 |
Hb_001040_150 |
0.1697319827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011853_010 |
0.170614399 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 20 |
Hb_177215_030 |
0.1706880605 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |