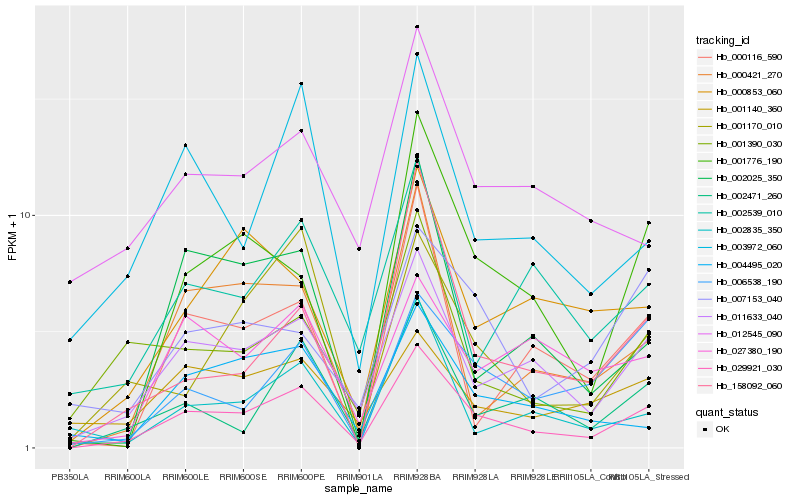
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029921_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011633_040 |
0.1293532098 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 3 |
Hb_002471_260 |
0.1783731769 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 4 |
Hb_003972_060 |
0.1873801857 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 5 |
Hb_001776_190 |
0.1873909122 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000421_270 |
0.1927691614 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 7 |
Hb_006538_190 |
0.1987887413 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001390_030 |
0.1988196418 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 9 |
Hb_027380_190 |
0.2000667536 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002025_350 |
0.2001717894 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 11 |
Hb_004495_020 |
0.200996044 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_002539_010 |
0.2016338525 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 13 |
Hb_001170_010 |
0.2034032621 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000853_060 |
0.205185914 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002835_350 |
0.2090469059 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 16 |
Hb_158092_060 |
0.2096849866 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_007153_040 |
0.2110206345 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001140_360 |
0.2131963805 |
- |
- |
- |
| 19 |
Hb_012545_090 |
0.2144648657 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |
| 20 |
Hb_000116_590 |
0.2192150729 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |