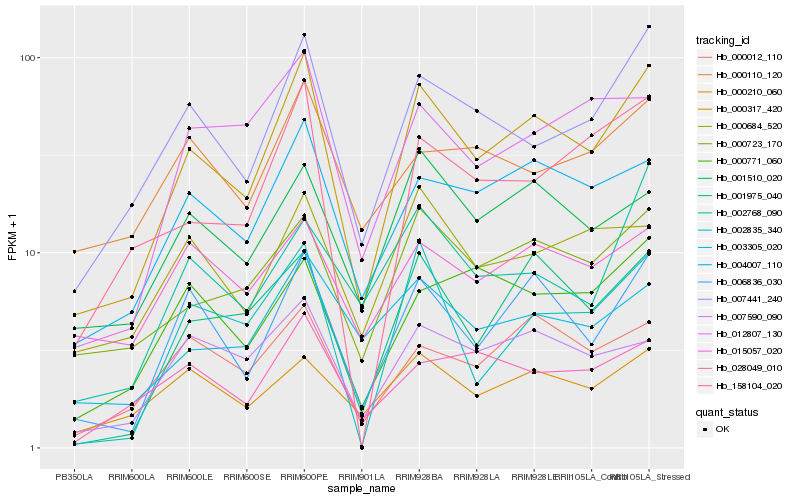
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_420 |
0.0 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 2 |
Hb_006836_030 |
0.1097887847 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 3 |
Hb_007441_240 |
0.12087068 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 4 |
Hb_004007_110 |
0.1281601883 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 5 |
Hb_000723_170 |
0.1313679004 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 6 |
Hb_001975_040 |
0.1369431177 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 7 |
Hb_158104_020 |
0.1484418496 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 8 |
Hb_001510_020 |
0.1494575185 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 9 |
Hb_000012_110 |
0.1507312672 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 10 |
Hb_007590_090 |
0.1516884661 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 11 |
Hb_000210_060 |
0.1569780896 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 12 |
Hb_003305_020 |
0.1575015938 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 13 |
Hb_002835_340 |
0.1597588341 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 14 |
Hb_000684_520 |
0.1607135721 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 15 |
Hb_012807_130 |
0.1613032293 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 16 |
Hb_000771_060 |
0.1641620606 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 17 |
Hb_028049_010 |
0.1647624656 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_015057_020 |
0.1703506666 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 19 |
Hb_002768_090 |
0.1707335959 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 20 |
Hb_000110_120 |
0.174998351 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |