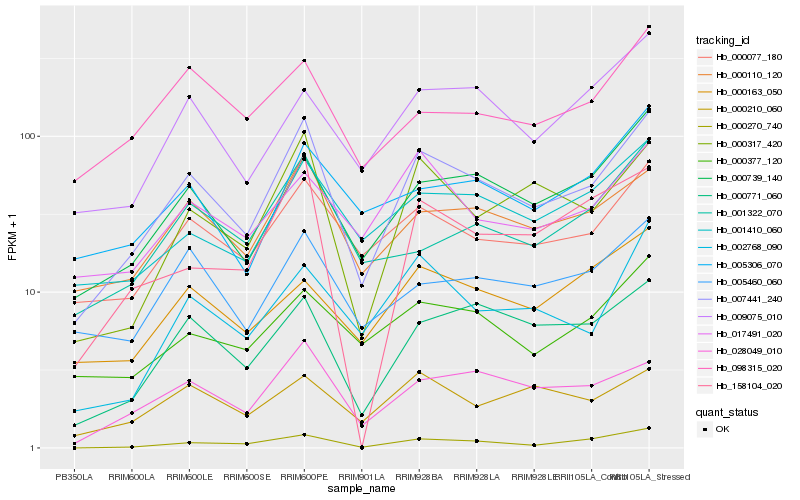
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_240 |
0.0 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 2 |
Hb_000110_120 |
0.1193316702 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 3 |
Hb_000077_180 |
0.119551277 |
- |
- |
- |
| 4 |
Hb_000739_140 |
0.1207057238 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 5 |
Hb_000317_420 |
0.12087068 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 6 |
Hb_028049_010 |
0.1239335025 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000771_060 |
0.1292470008 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 8 |
Hb_001410_060 |
0.1347014309 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 9 |
Hb_000270_740 |
0.1386061001 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 10 |
Hb_005460_060 |
0.14202163 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 11 |
Hb_000377_120 |
0.1455654928 |
- |
- |
PREDICTED: acyl carrier protein 3, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_002768_090 |
0.1463305815 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 13 |
Hb_009075_010 |
0.1475779598 |
- |
- |
60S ribosomal protein L10B [Hevea brasiliensis] |
| 14 |
Hb_001322_070 |
0.150030949 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_017491_020 |
0.1500898874 |
- |
- |
hypothetical protein B456_011G056700 [Gossypium raimondii] |
| 16 |
Hb_158104_020 |
0.1514926569 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 17 |
Hb_000210_060 |
0.1530409444 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 18 |
Hb_005306_070 |
0.1531644649 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 19 |
Hb_098315_020 |
0.1556261485 |
- |
- |
hypothetical protein M569_15712 [Genlisea aurea] |
| 20 |
Hb_000163_050 |
0.1564927307 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |