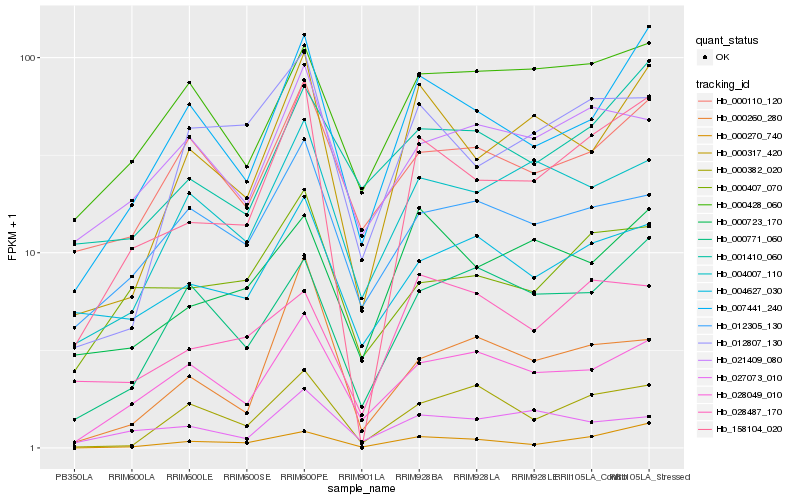
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158104_020 |
0.0 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 2 |
Hb_000317_420 |
0.1484418496 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 3 |
Hb_007441_240 |
0.1514926569 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 4 |
Hb_000270_740 |
0.1715894691 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 5 |
Hb_000407_070 |
0.1784125168 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 6 |
Hb_028049_010 |
0.1801283594 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000723_170 |
0.1805793402 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 8 |
Hb_004007_110 |
0.1857974281 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 9 |
Hb_000771_060 |
0.1921137694 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 10 |
Hb_012807_130 |
0.1923798283 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 11 |
Hb_001410_060 |
0.1967668439 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 12 |
Hb_027073_010 |
0.1967674301 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 13 |
Hb_004627_030 |
0.1970282903 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 14 |
Hb_028487_170 |
0.1977223777 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 15 |
Hb_000382_020 |
0.1979920717 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 16 |
Hb_000110_120 |
0.1983233589 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 17 |
Hb_000260_280 |
0.1992050868 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 18 |
Hb_012305_130 |
0.2006204151 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 19 |
Hb_000428_060 |
0.2016399478 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_021409_080 |
0.2025545671 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |