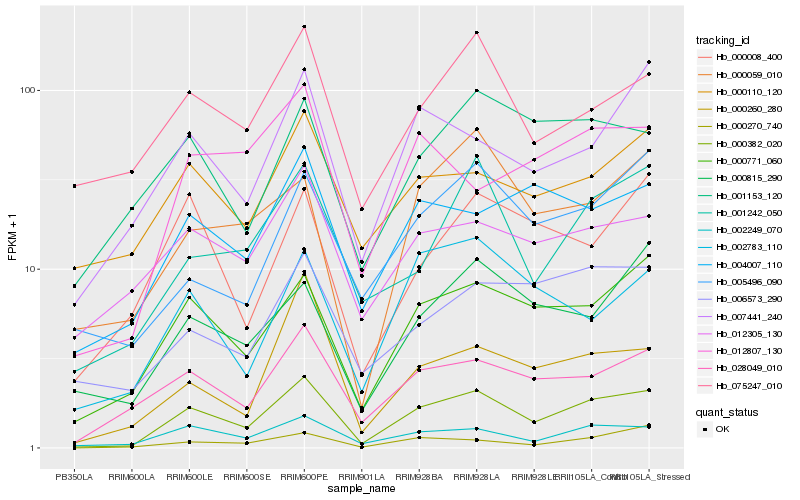
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000382_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 2 |
Hb_000771_060 |
0.132153837 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 3 |
Hb_002249_070 |
0.1388509103 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 4 |
Hb_028049_010 |
0.1481463997 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001242_050 |
0.1701833593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000270_740 |
0.1724414813 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 7 |
Hb_007441_240 |
0.1736348137 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 8 |
Hb_004007_110 |
0.1767032922 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 9 |
Hb_002783_110 |
0.1795028645 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 10 |
Hb_000815_290 |
0.1798519391 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 11 |
Hb_012807_130 |
0.1809534649 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 12 |
Hb_000059_010 |
0.1814108056 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000260_280 |
0.1819299994 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 14 |
Hb_075247_010 |
0.1830858536 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 15 |
Hb_005496_090 |
0.1832663879 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_012305_130 |
0.1835433289 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 17 |
Hb_000110_120 |
0.1847678817 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 18 |
Hb_006573_290 |
0.1863210071 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 19 |
Hb_000008_400 |
0.1875723298 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 20 |
Hb_001153_120 |
0.1898523806 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |