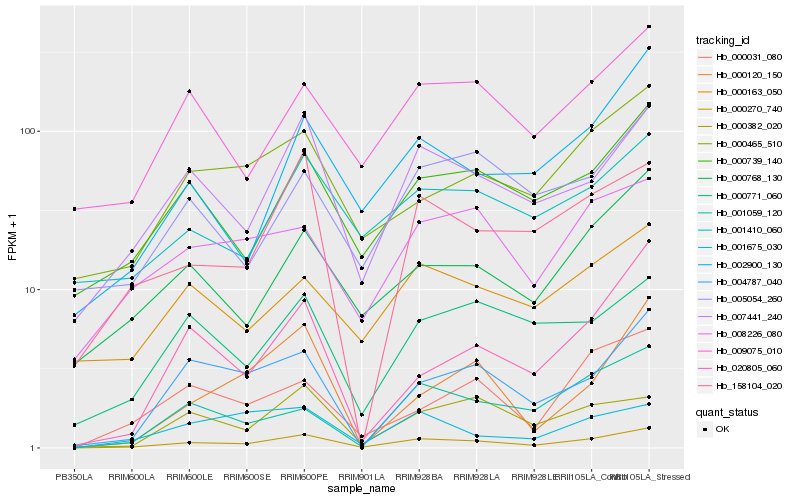
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_740 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 2 |
Hb_007441_240 |
0.1386061001 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 3 |
Hb_004787_040 |
0.1547649498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000739_140 |
0.1651819308 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 5 |
Hb_000768_130 |
0.1695126954 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 6 |
Hb_158104_020 |
0.1715894691 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 7 |
Hb_000382_020 |
0.1724414813 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 8 |
Hb_009075_010 |
0.1752466498 |
- |
- |
60S ribosomal protein L10B [Hevea brasiliensis] |
| 9 |
Hb_002900_130 |
0.1757575576 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 10 |
Hb_020805_060 |
0.1776022505 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 11 |
Hb_001059_120 |
0.177681902 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000465_510 |
0.1780903345 |
- |
- |
PREDICTED: GTP-binding protein YPTM2 [Populus euphratica] |
| 13 |
Hb_000771_060 |
0.178646973 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 14 |
Hb_000031_080 |
0.1824854458 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 15 |
Hb_008226_080 |
0.1868135277 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 16 |
Hb_005054_260 |
0.1876074671 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 17 |
Hb_000120_150 |
0.1887410986 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 18 |
Hb_001675_030 |
0.188896237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001410_060 |
0.1895217576 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 20 |
Hb_000163_050 |
0.1907872227 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |