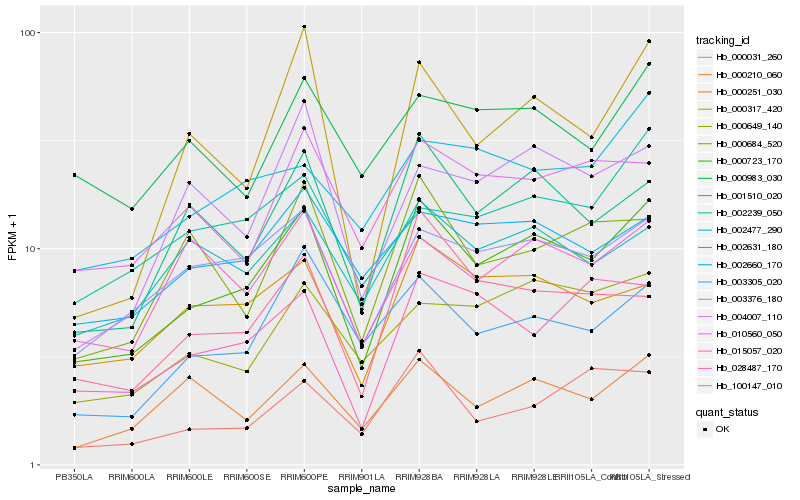
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_170 |
0.0 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 2 |
Hb_001510_020 |
0.1087575857 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 3 |
Hb_003376_180 |
0.1120535665 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 4 |
Hb_000251_030 |
0.1213873965 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_003305_020 |
0.1287302995 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 6 |
Hb_000210_060 |
0.1289384225 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 7 |
Hb_000317_420 |
0.1313679004 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 8 |
Hb_000649_140 |
0.1325838267 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002477_290 |
0.1340095441 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_010560_050 |
0.1346116623 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 11 |
Hb_002660_170 |
0.1349487037 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_028487_170 |
0.1362057078 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 13 |
Hb_100147_010 |
0.136442808 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 14 |
Hb_015057_020 |
0.1365146609 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 15 |
Hb_004007_110 |
0.139063233 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 16 |
Hb_000684_520 |
0.1395809481 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 17 |
Hb_002631_180 |
0.141137835 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 18 |
Hb_002239_050 |
0.1413731766 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000983_030 |
0.1418171153 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 20 |
Hb_000031_260 |
0.1419301151 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |