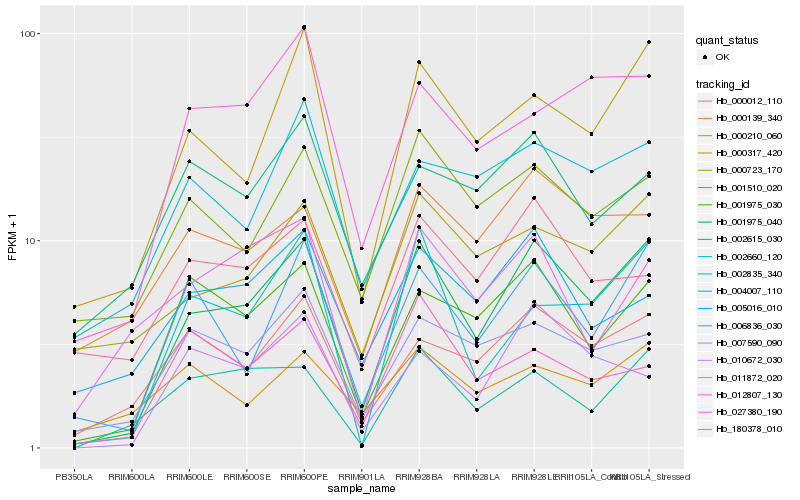
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001975_040 |
0.0 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 2 |
Hb_000317_420 |
0.1369431177 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 3 |
Hb_006836_030 |
0.148475551 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 4 |
Hb_001975_030 |
0.1508509971 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 5 |
Hb_002835_340 |
0.1582746946 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 6 |
Hb_000012_110 |
0.1586128182 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 7 |
Hb_002660_120 |
0.1645794115 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 8 |
Hb_000723_170 |
0.1652173816 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 9 |
Hb_007590_090 |
0.1663196396 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 10 |
Hb_012807_130 |
0.1722101187 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 11 |
Hb_001510_020 |
0.1735971541 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 12 |
Hb_004007_110 |
0.1762022661 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 13 |
Hb_000139_340 |
0.1793884935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010672_030 |
0.1849364383 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 15 |
Hb_005016_010 |
0.1898969721 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 16 |
Hb_180378_010 |
0.1939432819 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 17 |
Hb_000210_060 |
0.1947689564 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 18 |
Hb_002615_030 |
0.1959994593 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 19 |
Hb_011872_020 |
0.1962551331 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 20 |
Hb_027380_190 |
0.1967098239 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |