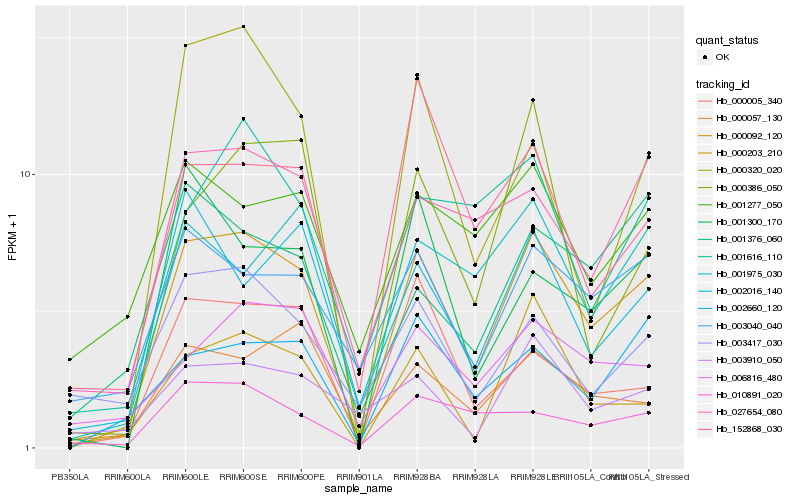
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000092_120 |
0.0 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |
| 2 |
Hb_027654_080 |
0.1602529518 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_003040_040 |
0.1652155482 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_002660_120 |
0.1653265966 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 5 |
Hb_010891_020 |
0.1685834705 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 6 |
Hb_000057_130 |
0.1687439407 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 7 |
Hb_002016_140 |
0.1691518695 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 8 |
Hb_000203_210 |
0.1702436968 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 9 |
Hb_001975_030 |
0.1719844424 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 10 |
Hb_001300_170 |
0.1765955098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006816_480 |
0.1768994873 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000005_340 |
0.179728993 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001376_060 |
0.1803262316 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC isoform X2 [Jatropha curcas] |
| 14 |
Hb_152868_030 |
0.1815092149 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 15 |
Hb_000386_050 |
0.1816071608 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_001616_110 |
0.1818643231 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 17 |
Hb_003417_030 |
0.1845481399 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000320_020 |
0.1845796498 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 19 |
Hb_001277_050 |
0.1883850767 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003910_050 |
0.1888862621 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |