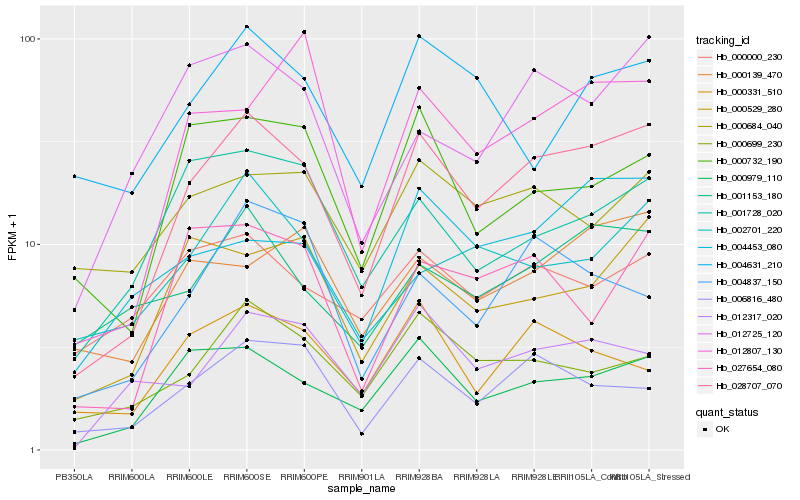
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028707_070 |
0.0 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_000979_110 |
0.1348849507 |
- |
- |
PREDICTED: GPI mannosyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_001153_180 |
0.1643089759 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 4 |
Hb_006816_480 |
0.1649385341 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004453_080 |
0.1661075004 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 6 |
Hb_012725_120 |
0.1661251757 |
- |
- |
PREDICTED: stress enhanced protein 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000699_230 |
0.1683183165 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 8 |
Hb_000529_280 |
0.1687052017 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000139_470 |
0.1717329289 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 10 |
Hb_027654_080 |
0.172506626 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_012317_020 |
0.174091084 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_000732_190 |
0.175410393 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 13 |
Hb_001728_020 |
0.1773623977 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 14 |
Hb_002701_220 |
0.1781002347 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 15 |
Hb_004631_210 |
0.1798154964 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 16 |
Hb_012807_130 |
0.1814164054 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 17 |
Hb_000331_510 |
0.182055675 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 18 |
Hb_000000_230 |
0.1828253564 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004837_150 |
0.1833895867 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 20 |
Hb_000684_040 |
0.18340135 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |