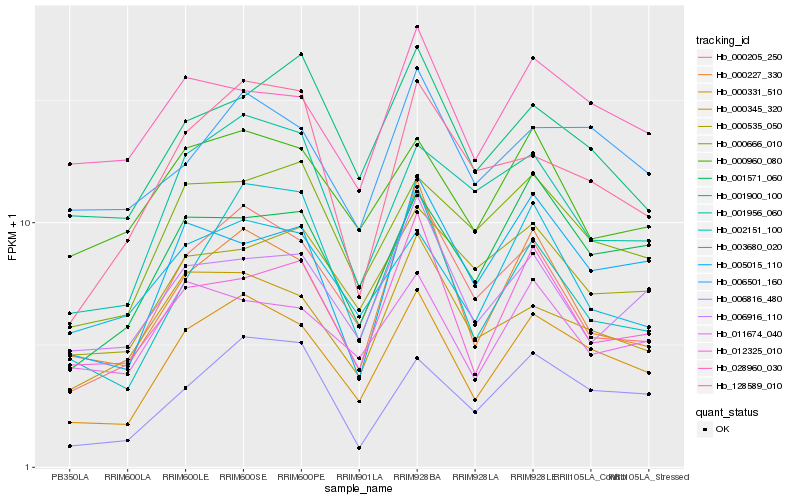
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_510 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 2 |
Hb_006816_480 |
0.1110704708 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001571_060 |
0.1111429033 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 4 |
Hb_000205_250 |
0.1136368933 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 5 |
Hb_000345_320 |
0.1163210992 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000666_010 |
0.1177737037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000227_330 |
0.1189065588 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 8 |
Hb_006501_160 |
0.1218155408 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 9 |
Hb_012325_010 |
0.1220109418 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 10 |
Hb_005015_110 |
0.1239280047 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 11 |
Hb_001956_060 |
0.1239561357 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 12 |
Hb_011674_040 |
0.129084578 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003680_020 |
0.1301578829 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000960_080 |
0.1303052839 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_128589_010 |
0.1305229763 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 16 |
Hb_002151_100 |
0.131421537 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 17 |
Hb_000535_050 |
0.1320646493 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 18 |
Hb_028960_030 |
0.1325323283 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 19 |
Hb_001900_100 |
0.1327203108 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006916_110 |
0.1340371254 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |