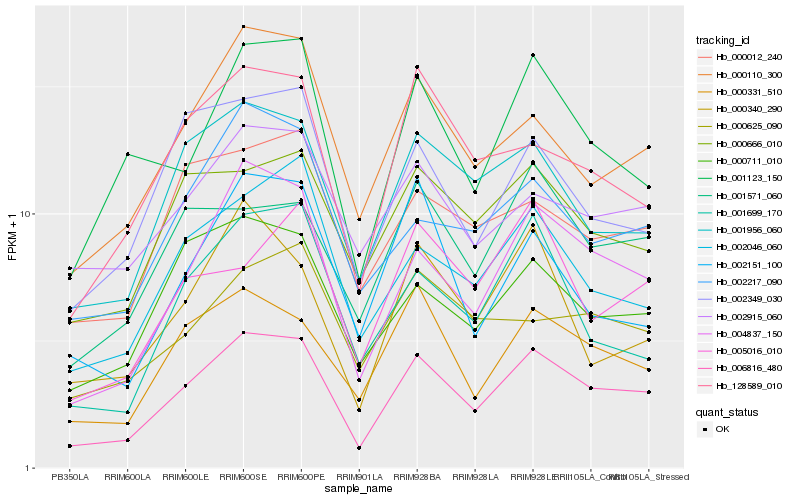
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_480 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004837_150 |
0.0807184883 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 3 |
Hb_000331_510 |
0.1110704708 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 4 |
Hb_000110_300 |
0.1122442708 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 5 |
Hb_001956_060 |
0.1150195143 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 6 |
Hb_001123_150 |
0.1190003893 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 7 |
Hb_002046_060 |
0.1206786478 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 8 |
Hb_000711_010 |
0.1230397256 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 9 |
Hb_128589_010 |
0.1241553117 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 10 |
Hb_000666_010 |
0.1252355534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001571_060 |
0.1260716661 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 12 |
Hb_002217_090 |
0.1289033398 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 13 |
Hb_002349_030 |
0.1303838341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000012_240 |
0.1319064093 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001699_170 |
0.132050169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000340_290 |
0.1328159205 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |
| 17 |
Hb_002915_060 |
0.1368174852 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 18 |
Hb_005016_010 |
0.1385647233 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 19 |
Hb_000625_090 |
0.1395210656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002151_100 |
0.1405042303 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |