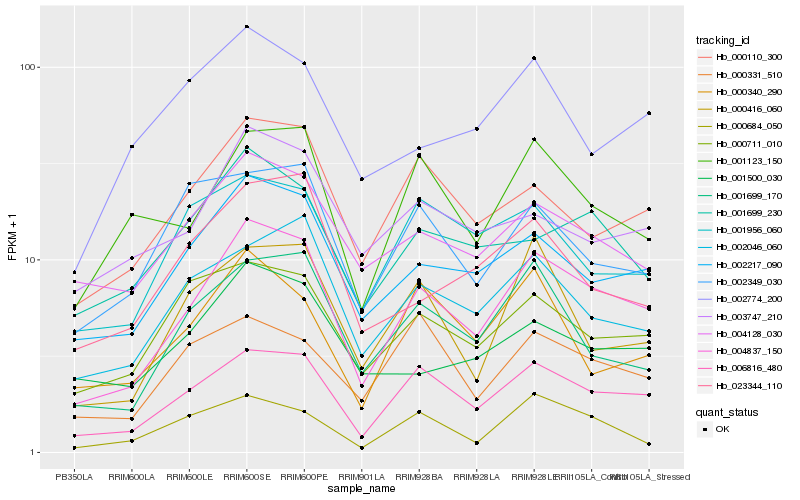
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004837_150 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 2 |
Hb_006816_480 |
0.0807184883 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002217_090 |
0.1135822247 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 4 |
Hb_001699_170 |
0.1346850131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000110_300 |
0.1357202138 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 6 |
Hb_002046_060 |
0.1364333852 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 7 |
Hb_001123_150 |
0.1387053953 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 8 |
Hb_000711_010 |
0.1415697489 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 9 |
Hb_004128_030 |
0.1415972776 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000416_060 |
0.1490499247 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 11 |
Hb_001699_230 |
0.1506473575 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 12 |
Hb_023344_110 |
0.1558591556 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 13 |
Hb_001500_030 |
0.1562115788 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001956_060 |
0.1567888831 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 15 |
Hb_003747_210 |
0.1582226146 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 16 |
Hb_000331_510 |
0.1594786815 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 17 |
Hb_000340_290 |
0.1604633673 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |
| 18 |
Hb_002349_030 |
0.1607256322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002774_200 |
0.1610394877 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 20 |
Hb_000684_050 |
0.1628609021 |
- |
- |
PREDICTED: MLO-like protein 6 [Sesamum indicum] |