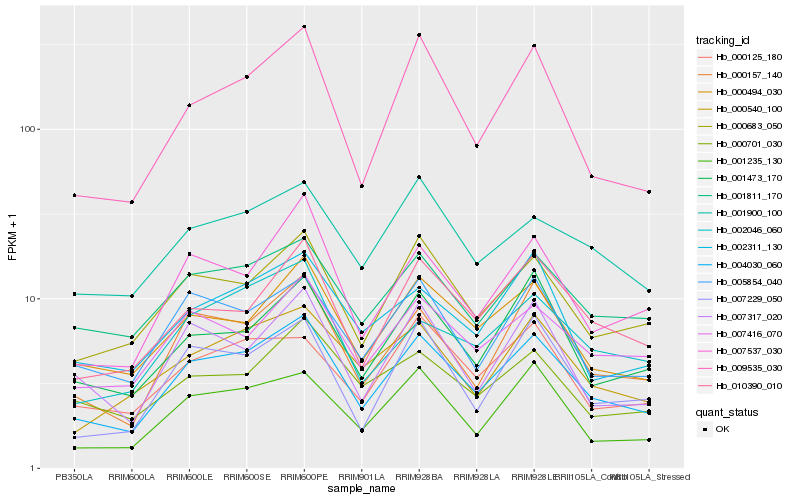
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004030_060 |
0.0 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 2 |
Hb_000157_140 |
0.0803891558 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 3 |
Hb_000683_050 |
0.0811474776 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 4 |
Hb_000494_030 |
0.0835456674 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 5 |
Hb_009535_030 |
0.0879752368 |
- |
- |
CP2 [Hevea brasiliensis] |
| 6 |
Hb_010390_010 |
0.0893808737 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 7 |
Hb_005854_040 |
0.0926129993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001811_170 |
0.0934040254 |
- |
- |
dynamin, putative [Ricinus communis] |
| 9 |
Hb_001473_170 |
0.0949595337 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 10 |
Hb_007537_030 |
0.0954391493 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007416_070 |
0.0959070924 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 12 |
Hb_001235_130 |
0.096477157 |
- |
- |
- |
| 13 |
Hb_001900_100 |
0.1022306106 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000540_100 |
0.1025385055 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 15 |
Hb_002311_130 |
0.1033422375 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002046_060 |
0.1056305069 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 17 |
Hb_007229_050 |
0.1062017183 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007317_020 |
0.1063091104 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000125_180 |
0.1086268939 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 20 |
Hb_000701_030 |
0.109091376 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |