| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
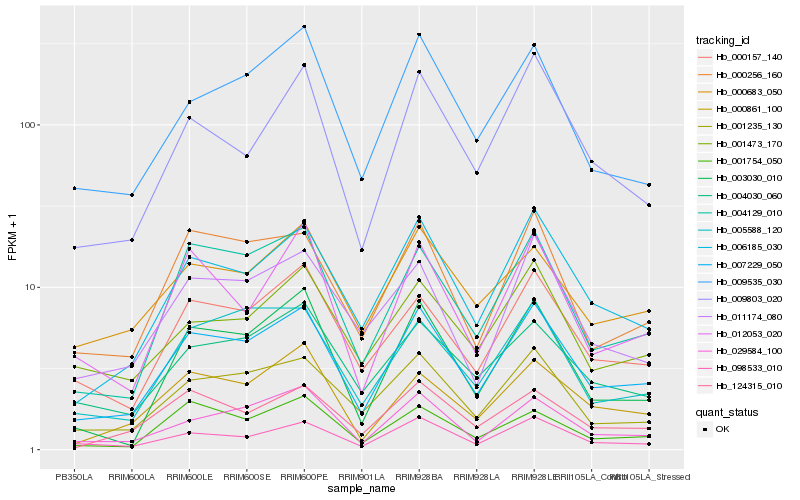
Hb_007229_050 |
0.0 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006185_030 |
0.076401068 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 3 |
Hb_004129_010 |
0.083217975 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 4 |
Hb_001235_130 |
0.0857820482 |
- |
- |
- |
| 5 |
Hb_098533_010 |
0.0889124442 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 6 |
Hb_000157_140 |
0.0903546011 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_000256_160 |
0.0920205024 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 8 |
Hb_029584_100 |
0.0963102436 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 9 |
Hb_009535_030 |
0.1019602654 |
- |
- |
CP2 [Hevea brasiliensis] |
| 10 |
Hb_009803_020 |
0.1055976403 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 11 |
Hb_004030_060 |
0.1062017183 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 12 |
Hb_011174_080 |
0.1064147817 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 13 |
Hb_000861_100 |
0.1067330882 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 14 |
Hb_124315_010 |
0.1080477177 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |
| 15 |
Hb_005588_120 |
0.1081770033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003030_010 |
0.1087697385 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 17 |
Hb_000683_050 |
0.11137642 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 18 |
Hb_012053_020 |
0.1115949405 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001473_170 |
0.1116690516 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 20 |
Hb_001754_050 |
0.1137245632 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |