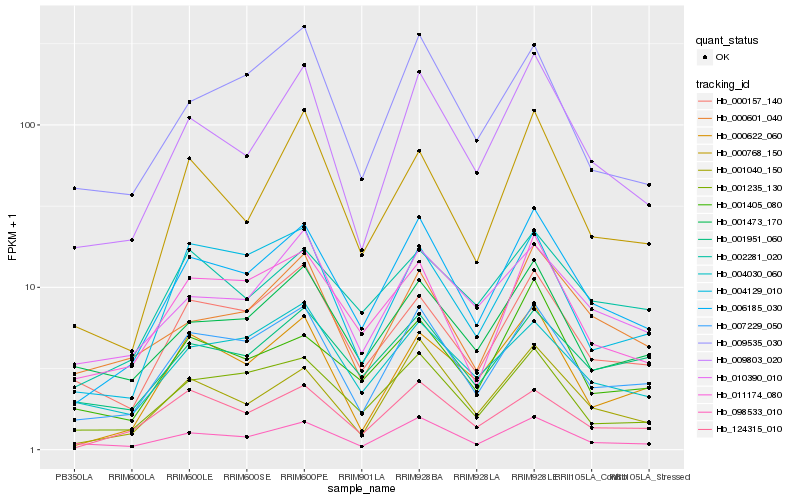
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006185_030 |
0.0 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 2 |
Hb_007229_050 |
0.076401068 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009803_020 |
0.0910427631 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 4 |
Hb_001040_150 |
0.0945148571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001235_130 |
0.097530656 |
- |
- |
- |
| 6 |
Hb_098533_010 |
0.1058778863 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 7 |
Hb_011174_080 |
0.1062911679 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 8 |
Hb_124315_010 |
0.1086560322 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |
| 9 |
Hb_000157_140 |
0.1176825452 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_002281_020 |
0.1222444756 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 11 |
Hb_001473_170 |
0.1222737276 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 12 |
Hb_010390_010 |
0.1236978924 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 13 |
Hb_009535_030 |
0.1244025448 |
- |
- |
CP2 [Hevea brasiliensis] |
| 14 |
Hb_001951_060 |
0.1247661741 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 15 |
Hb_000601_040 |
0.1254544849 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 16 |
Hb_000768_150 |
0.1269998349 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 17 |
Hb_001405_080 |
0.1277599353 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 18 |
Hb_004030_060 |
0.1282029317 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 19 |
Hb_004129_010 |
0.1292724393 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 20 |
Hb_000622_060 |
0.1308351755 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |