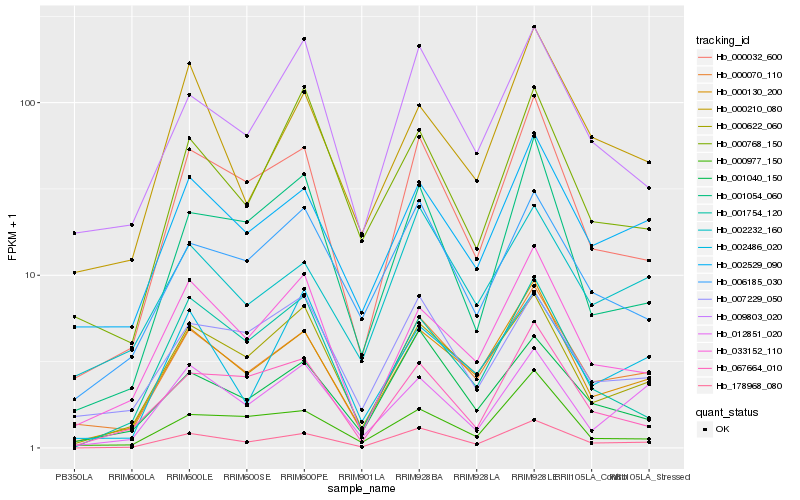
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178968_080 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000130_200 |
0.0978033162 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 3 |
Hb_000032_600 |
0.0984157513 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 4 |
Hb_000070_110 |
0.1044155269 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001040_150 |
0.1228672691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000622_060 |
0.1287131885 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001054_060 |
0.1349212627 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 8 |
Hb_002529_090 |
0.1456566837 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_067664_010 |
0.1476915309 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 10 |
Hb_033152_110 |
0.1487527686 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 11 |
Hb_006185_030 |
0.1515561455 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 12 |
Hb_009803_020 |
0.1520439267 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 13 |
Hb_000977_150 |
0.1554727656 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 14 |
Hb_002232_160 |
0.1568766373 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 15 |
Hb_001754_120 |
0.1570160387 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 16 |
Hb_012851_020 |
0.1586291999 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_000210_080 |
0.1630220608 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_007229_050 |
0.1685838231 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000768_150 |
0.1686412679 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 20 |
Hb_002486_020 |
0.1687272149 |
- |
- |
ATP binding protein, putative [Ricinus communis] |