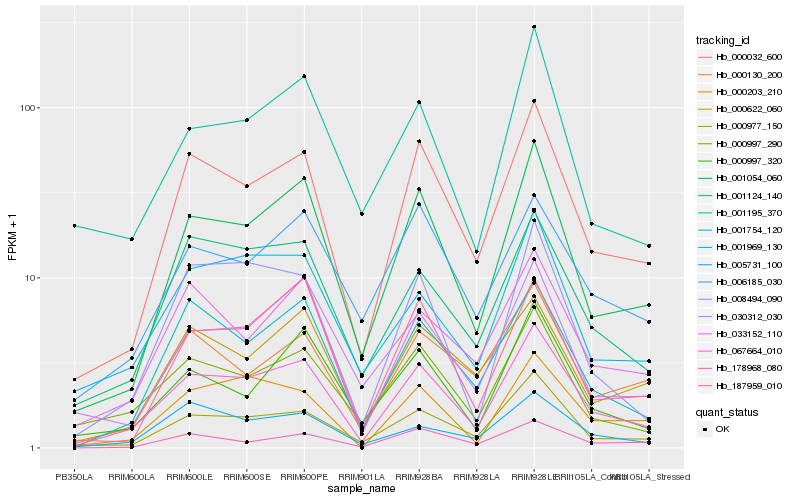
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_067664_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 2 |
Hb_000032_600 |
0.0804184791 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 3 |
Hb_001054_060 |
0.0902518122 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 4 |
Hb_000977_150 |
0.1036411341 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 5 |
Hb_001754_120 |
0.1169163426 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 6 |
Hb_000997_290 |
0.1217502249 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 7 |
Hb_001124_140 |
0.1223558732 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 8 |
Hb_033152_110 |
0.1250710686 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 9 |
Hb_008494_090 |
0.1296114116 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 10 |
Hb_001969_130 |
0.1318642888 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 11 |
Hb_000203_210 |
0.1354437696 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 12 |
Hb_000997_320 |
0.1375616445 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 13 |
Hb_187959_010 |
0.1392334706 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 14 |
Hb_005731_100 |
0.1396259369 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 15 |
Hb_001195_370 |
0.1429345295 |
- |
- |
- |
| 16 |
Hb_000622_060 |
0.1432367036 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000130_200 |
0.1461694793 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 18 |
Hb_030312_030 |
0.1470610298 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006185_030 |
0.1476311323 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 20 |
Hb_178968_080 |
0.1476915309 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |