| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
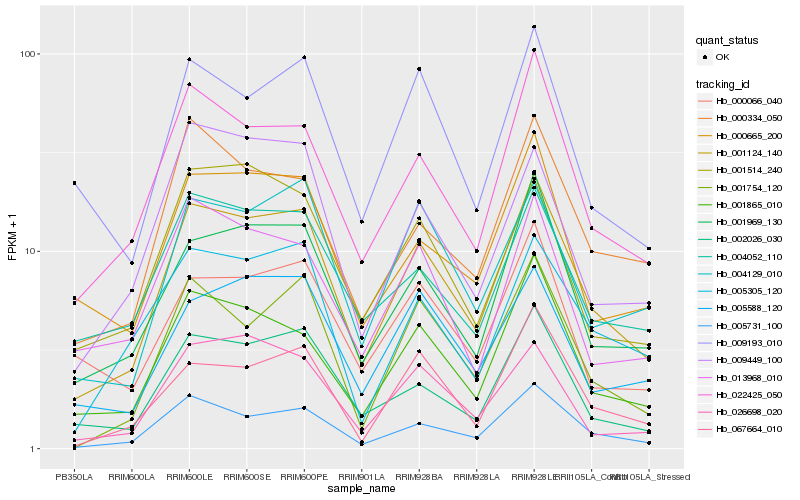
Hb_001124_140 |
0.0 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 2 |
Hb_005731_100 |
0.0781973792 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 3 |
Hb_001969_130 |
0.0929751699 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 4 |
Hb_022425_050 |
0.0937051041 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 5 |
Hb_002026_030 |
0.0959817802 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 6 |
Hb_001865_010 |
0.1007642115 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 7 |
Hb_004129_010 |
0.1113268429 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_000665_200 |
0.1138511066 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_001754_120 |
0.1149887013 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 10 |
Hb_004052_110 |
0.117262325 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005588_120 |
0.11824149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000334_050 |
0.1185033536 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 13 |
Hb_009193_010 |
0.1204084265 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 14 |
Hb_005305_120 |
0.1206378549 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 15 |
Hb_067664_010 |
0.1223558732 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 16 |
Hb_026698_020 |
0.124219295 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 17 |
Hb_001514_240 |
0.1264075013 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 18 |
Hb_013968_010 |
0.1265075182 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 19 |
Hb_009449_100 |
0.1266045159 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 20 |
Hb_000066_040 |
0.1273744867 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |