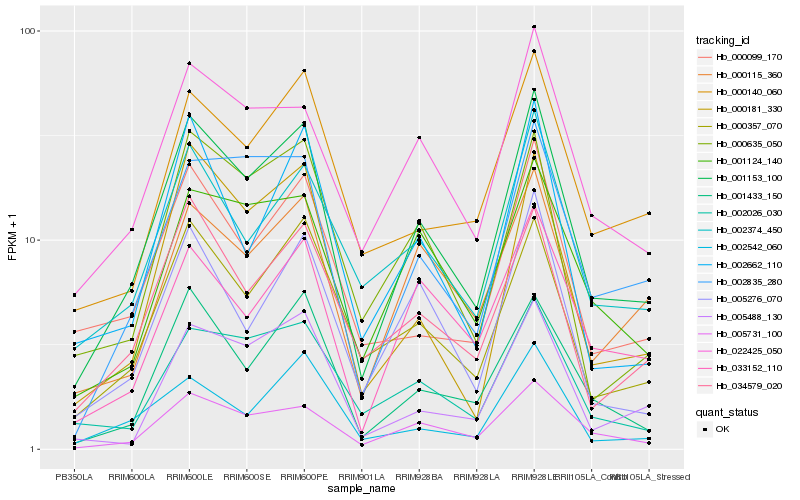
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001153_100 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 2 |
Hb_005731_100 |
0.0975298745 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 3 |
Hb_000357_070 |
0.1114021024 |
- |
- |
- |
| 4 |
Hb_001433_150 |
0.1155774473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002835_280 |
0.1171383024 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 6 |
Hb_022425_050 |
0.1200664725 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 7 |
Hb_034579_020 |
0.1208925862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002374_450 |
0.1253101101 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000115_360 |
0.1255529754 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000181_330 |
0.1266102138 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 11 |
Hb_002026_030 |
0.1278317546 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 12 |
Hb_005276_070 |
0.1282417874 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 13 |
Hb_000635_050 |
0.1289955221 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 14 |
Hb_001124_140 |
0.1300907928 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 15 |
Hb_000099_170 |
0.1303186945 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 16 |
Hb_033152_110 |
0.1306629435 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 17 |
Hb_000140_060 |
0.132193618 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 18 |
Hb_005488_130 |
0.1332504331 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_002662_110 |
0.1343849643 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 20 |
Hb_002542_060 |
0.1364038114 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |