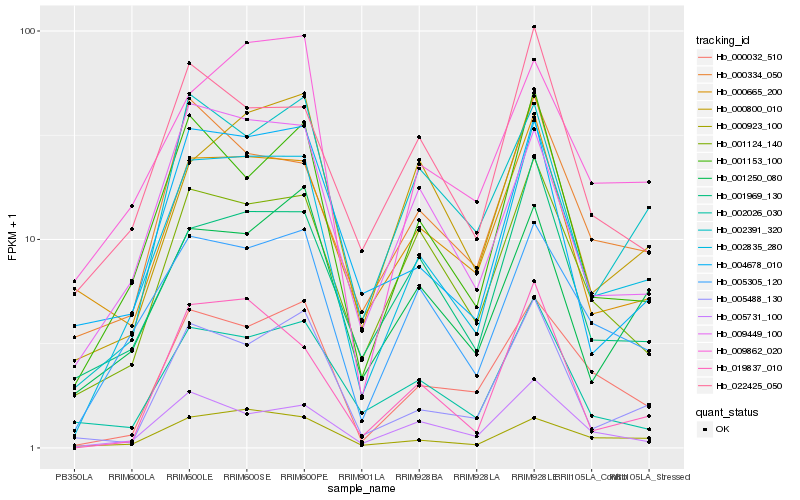
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_280 |
0.0 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 2 |
Hb_005305_120 |
0.1143667347 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 3 |
Hb_001153_100 |
0.1171383024 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 4 |
Hb_001969_130 |
0.1294603382 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 5 |
Hb_000032_510 |
0.1324710279 |
- |
- |
PREDICTED: SUN domain-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_000923_100 |
0.1335697653 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 7 |
Hb_001124_140 |
0.1341695991 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 8 |
Hb_005731_100 |
0.1360698299 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 9 |
Hb_005488_130 |
0.1371756331 |
- |
- |
transporter, putative [Ricinus communis] |
| 10 |
Hb_009449_100 |
0.1414869387 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 11 |
Hb_004678_010 |
0.1461542332 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 12 |
Hb_009862_020 |
0.148520663 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 13 |
Hb_002026_030 |
0.1492199529 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 14 |
Hb_000800_010 |
0.1495747405 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 15 |
Hb_001250_080 |
0.1501554646 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 16 |
Hb_000334_050 |
0.1516023994 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 17 |
Hb_022425_050 |
0.1521291721 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 18 |
Hb_000665_200 |
0.1530725132 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 19 |
Hb_002391_320 |
0.1544829673 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 20 |
Hb_019837_010 |
0.1550743824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |