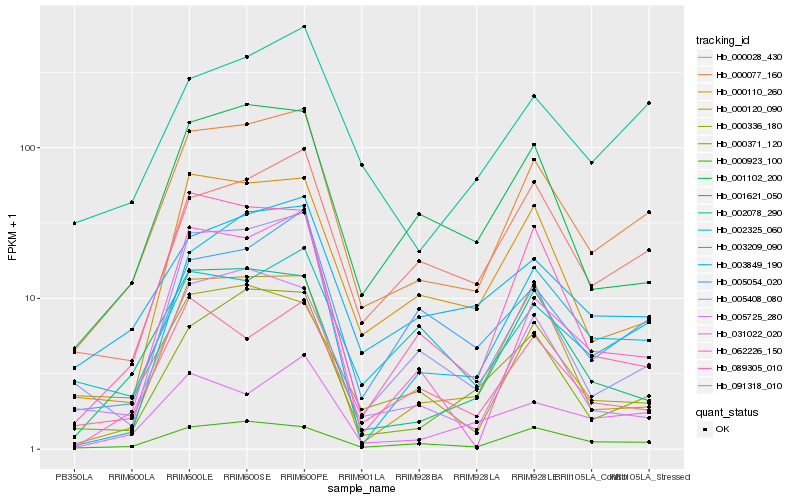
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_160 |
0.0 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 2 |
Hb_000923_100 |
0.1306295554 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 3 |
Hb_000371_120 |
0.1311305197 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_003849_190 |
0.1344381702 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 5 |
Hb_000028_430 |
0.1356668964 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000110_260 |
0.1367379957 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 7 |
Hb_002078_290 |
0.1384697481 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |
| 8 |
Hb_001621_050 |
0.1394722995 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 9 |
Hb_005408_080 |
0.139571525 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 10 |
Hb_001102_200 |
0.143064947 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 11 |
Hb_003209_090 |
0.1436185522 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 12 |
Hb_005054_020 |
0.1457014628 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 13 |
Hb_031022_020 |
0.1528927467 |
- |
- |
- |
| 14 |
Hb_091318_010 |
0.1536191731 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 15 |
Hb_000336_180 |
0.1551889769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002325_060 |
0.1570029845 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 17 |
Hb_089305_010 |
0.1571480987 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 18 |
Hb_005725_280 |
0.1584727635 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 19 |
Hb_062226_150 |
0.1625744558 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000120_090 |
0.1627752303 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |