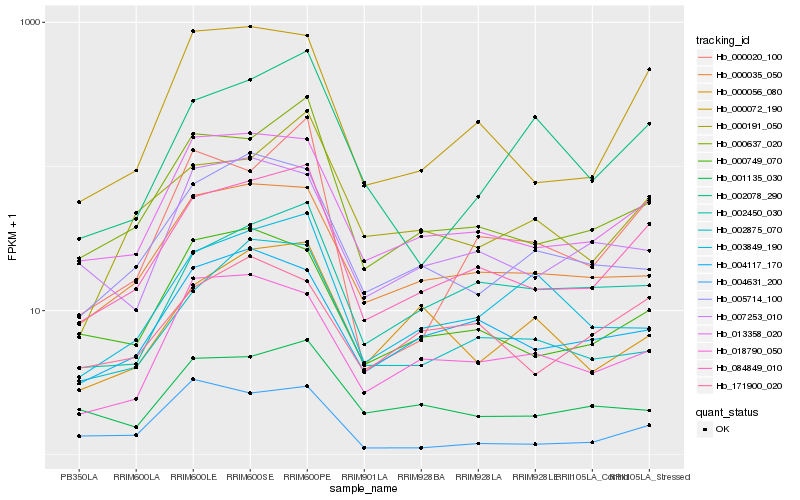
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084849_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_013358_020 |
0.0980797606 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 3 |
Hb_000637_020 |
0.0986500207 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 4 |
Hb_000072_190 |
0.1127932432 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 5 |
Hb_000035_050 |
0.1225342948 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 6 |
Hb_002450_030 |
0.1320957817 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 7 |
Hb_000749_070 |
0.1372438513 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 8 |
Hb_004117_170 |
0.137690403 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 9 |
Hb_000191_050 |
0.1397240298 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 10 |
Hb_002875_070 |
0.1407128477 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 11 |
Hb_000020_100 |
0.1464389337 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 12 |
Hb_018790_050 |
0.1518547362 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 13 |
Hb_001135_030 |
0.1522536896 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 14 |
Hb_005714_100 |
0.1523243443 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 15 |
Hb_171900_020 |
0.1525484374 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 16 |
Hb_007253_010 |
0.152853652 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 17 |
Hb_003849_190 |
0.1595844346 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 18 |
Hb_002078_290 |
0.1626285407 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |
| 19 |
Hb_000056_080 |
0.1644987019 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004631_200 |
0.1651976196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |