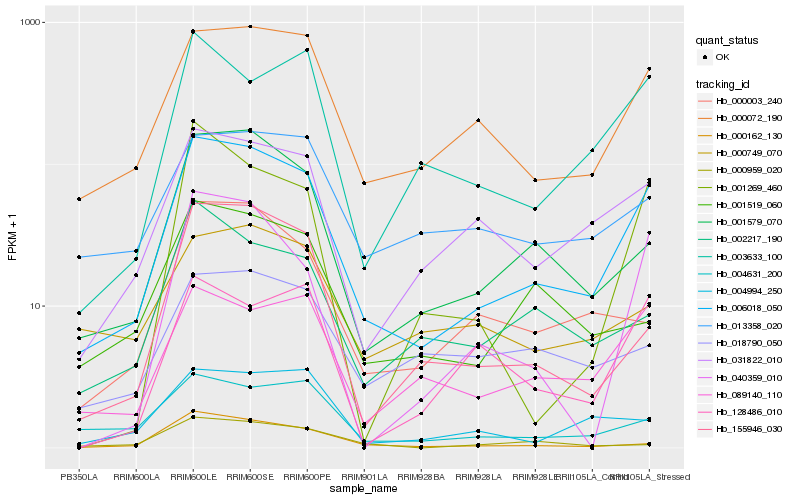
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006018_050 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 2 |
Hb_000072_190 |
0.1406580039 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 3 |
Hb_001579_070 |
0.1529159976 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 4 |
Hb_031822_010 |
0.1600234354 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 5 |
Hb_089140_110 |
0.1703647866 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000162_130 |
0.1791090553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003633_100 |
0.1796583854 |
- |
- |
hypoia-responsive family protein 1 [Hevea brasiliensis] |
| 8 |
Hb_004631_200 |
0.1811126512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_155946_030 |
0.1842682767 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 10 |
Hb_000003_240 |
0.1854355177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002217_190 |
0.1910718587 |
- |
- |
invertase [Hevea brasiliensis] |
| 12 |
Hb_040359_010 |
0.1950465216 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 13 |
Hb_001519_060 |
0.1951344776 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_004994_250 |
0.1982420651 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 15 |
Hb_001269_460 |
0.2042854447 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 16 |
Hb_013358_020 |
0.2043116875 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 17 |
Hb_128486_010 |
0.2069961126 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 18 |
Hb_018790_050 |
0.2095848264 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 19 |
Hb_000959_020 |
0.2131650247 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_000749_070 |
0.2134278562 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |