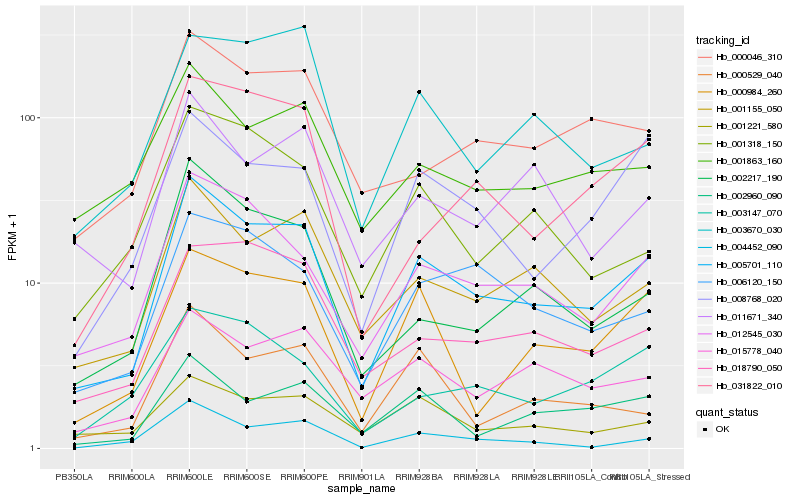
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005701_110 |
0.0 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 2 |
Hb_012545_030 |
0.1211231666 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 3 |
Hb_001155_050 |
0.1252514858 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 4 |
Hb_000984_260 |
0.1453467494 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 5 |
Hb_006120_150 |
0.146947646 |
- |
- |
invertase [Hevea brasiliensis] |
| 6 |
Hb_015778_040 |
0.1498132152 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 7 |
Hb_008768_020 |
0.1498307092 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 8 |
Hb_002960_090 |
0.1508765722 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000046_310 |
0.1532911499 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 10 |
Hb_001221_580 |
0.1540258577 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 11 |
Hb_018790_050 |
0.1540926214 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 12 |
Hb_002217_190 |
0.1567466289 |
- |
- |
invertase [Hevea brasiliensis] |
| 13 |
Hb_004452_090 |
0.1585835704 |
- |
- |
hypothetical protein JCGZ_12231 [Jatropha curcas] |
| 14 |
Hb_001863_160 |
0.1586404553 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 15 |
Hb_000529_040 |
0.1610819435 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 16 |
Hb_031822_010 |
0.1629705022 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 17 |
Hb_001318_150 |
0.1652001221 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 18 |
Hb_011671_340 |
0.165205029 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 19 |
Hb_003670_030 |
0.1674810604 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 20 |
Hb_003147_070 |
0.1676292043 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |