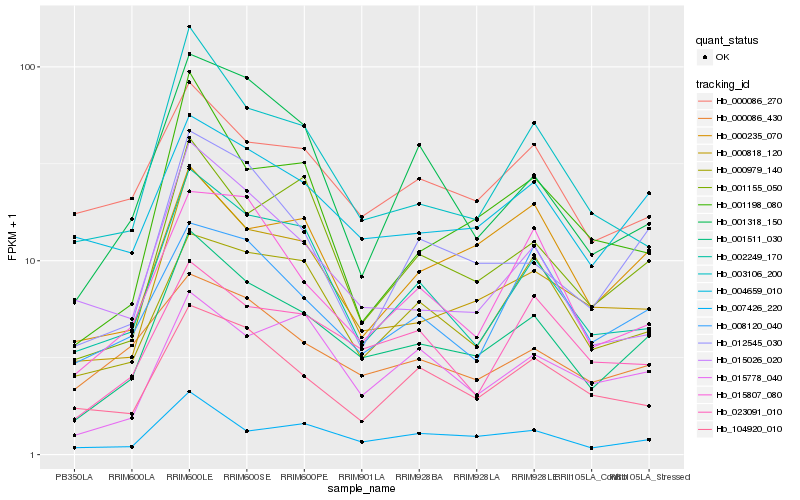
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001511_030 |
0.0 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 [Jatropha curcas] |
| 2 |
Hb_000818_120 |
0.1015596851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007426_220 |
0.1107615985 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002249_170 |
0.122572717 |
- |
- |
copine, putative [Ricinus communis] |
| 5 |
Hb_012545_030 |
0.1260426836 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 6 |
Hb_004659_010 |
0.1269217004 |
- |
- |
Guanine nucleotide-binding subunit beta-2 [Gossypium arboreum] |
| 7 |
Hb_001155_050 |
0.1297435385 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 8 |
Hb_003106_200 |
0.1362591556 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 9 |
Hb_023091_010 |
0.1384032142 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 10 |
Hb_000086_430 |
0.1410466591 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 11 |
Hb_000086_270 |
0.141736263 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 12 |
Hb_015778_040 |
0.1437899838 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 13 |
Hb_015807_080 |
0.1440381178 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 14 |
Hb_015026_020 |
0.1446366202 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 15 |
Hb_000235_070 |
0.1455568434 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 16 |
Hb_008120_040 |
0.146014667 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 17 |
Hb_104920_010 |
0.1467742362 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 18 |
Hb_000979_140 |
0.1472979056 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 19 |
Hb_001318_150 |
0.148095866 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 20 |
Hb_001198_080 |
0.1487513232 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |