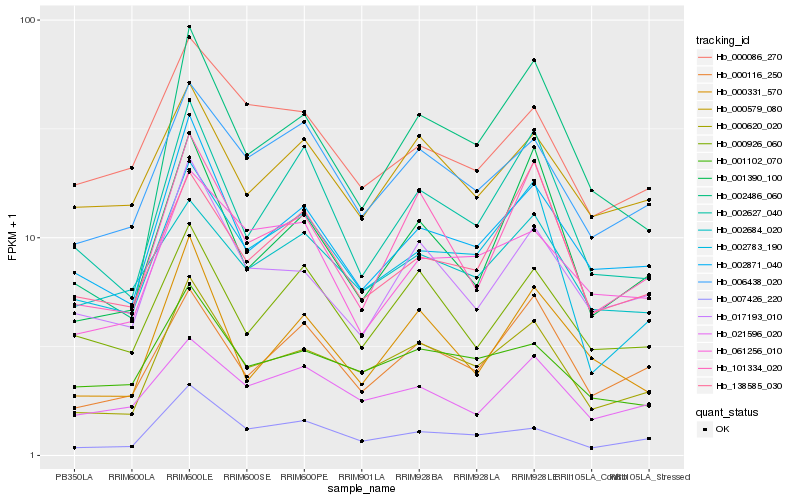
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000620_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649917 [Jatropha curcas] |
| 2 |
Hb_002871_040 |
0.0991297916 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 3 |
Hb_001102_070 |
0.1046970853 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_101334_020 |
0.1069560873 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 5 |
Hb_002783_190 |
0.107380769 |
- |
- |
PREDICTED: uncharacterized protein LOC105635339 [Jatropha curcas] |
| 6 |
Hb_006438_020 |
0.1078233878 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 7 |
Hb_007426_220 |
0.1089976237 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_002486_060 |
0.1099074218 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 9 |
Hb_017193_010 |
0.1128039619 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000116_250 |
0.1128492135 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 11 |
Hb_001390_100 |
0.1147493321 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002627_040 |
0.1203326163 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 13 |
Hb_021596_020 |
0.125006967 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 14 |
Hb_000926_060 |
0.1253727365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000579_080 |
0.1254720231 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 16 |
Hb_000331_570 |
0.1274111396 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 17 |
Hb_002684_020 |
0.1326882877 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 18 |
Hb_061256_010 |
0.133378906 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000086_270 |
0.1348848274 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 20 |
Hb_138585_030 |
0.1350884293 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |