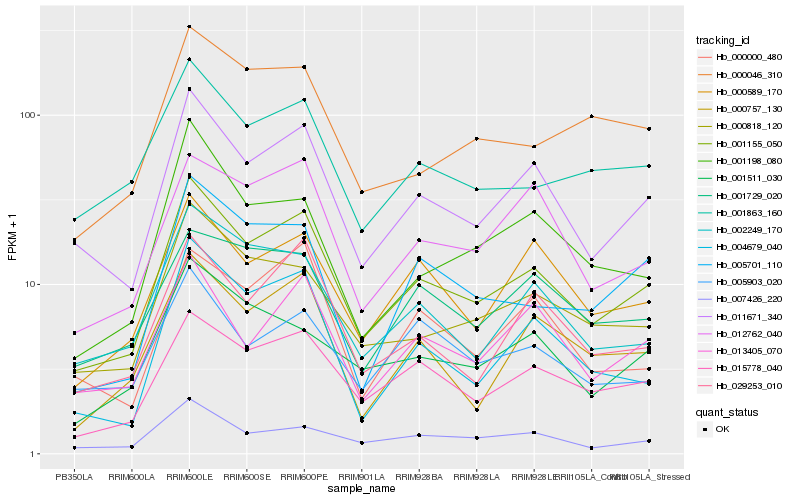
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001155_050 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 2 |
Hb_011671_340 |
0.0700814797 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 3 |
Hb_015778_040 |
0.0920534431 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 4 |
Hb_007426_220 |
0.109073065 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000818_120 |
0.1105408606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000589_170 |
0.1139172936 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_029253_010 |
0.1146348031 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 8 |
Hb_002249_170 |
0.1156663094 |
- |
- |
copine, putative [Ricinus communis] |
| 9 |
Hb_013405_070 |
0.1158564064 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 10 |
Hb_004679_040 |
0.1196618366 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 11 |
Hb_012762_040 |
0.1202443481 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 12 |
Hb_001863_160 |
0.121210587 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 13 |
Hb_000757_130 |
0.1216421947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005701_110 |
0.1252514858 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 15 |
Hb_001198_080 |
0.1289503488 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001511_030 |
0.1297435385 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 [Jatropha curcas] |
| 17 |
Hb_000000_480 |
0.1315464989 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001729_020 |
0.1319641823 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000046_310 |
0.1329674149 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 20 |
Hb_005903_020 |
0.1343459731 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |