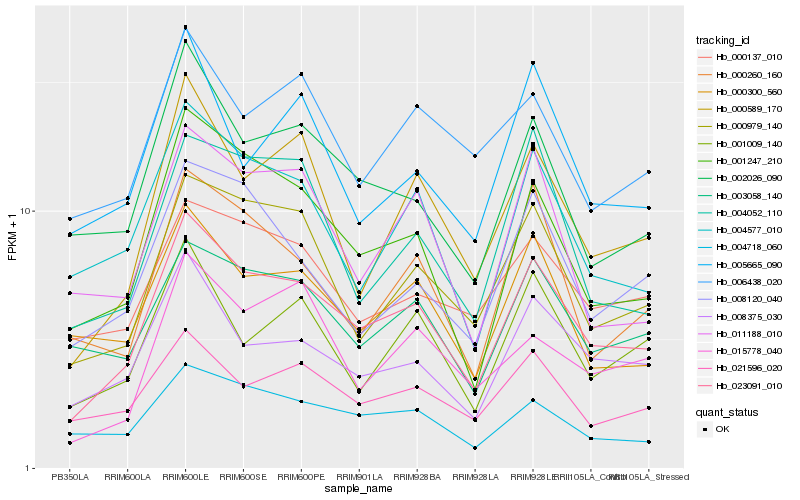
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023091_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 2 |
Hb_001247_210 |
0.0978963002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008375_030 |
0.1047353556 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 4 |
Hb_000979_140 |
0.1054512908 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 5 |
Hb_000589_170 |
0.1078784405 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 6 |
Hb_004577_010 |
0.1126737198 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 7 |
Hb_002026_090 |
0.1128481668 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 8 |
Hb_004718_060 |
0.1152362839 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_021596_020 |
0.117292592 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 10 |
Hb_003058_140 |
0.1173982496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000300_560 |
0.1187018148 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000137_010 |
0.1217657721 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 13 |
Hb_001009_140 |
0.1228081307 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 14 |
Hb_008120_040 |
0.1228093573 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 15 |
Hb_000260_160 |
0.1257181331 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_011188_010 |
0.1271324708 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 17 |
Hb_015778_040 |
0.1299500551 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 18 |
Hb_006438_020 |
0.1308259108 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 19 |
Hb_005665_090 |
0.1314383853 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 20 |
Hb_004052_110 |
0.1320511809 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |