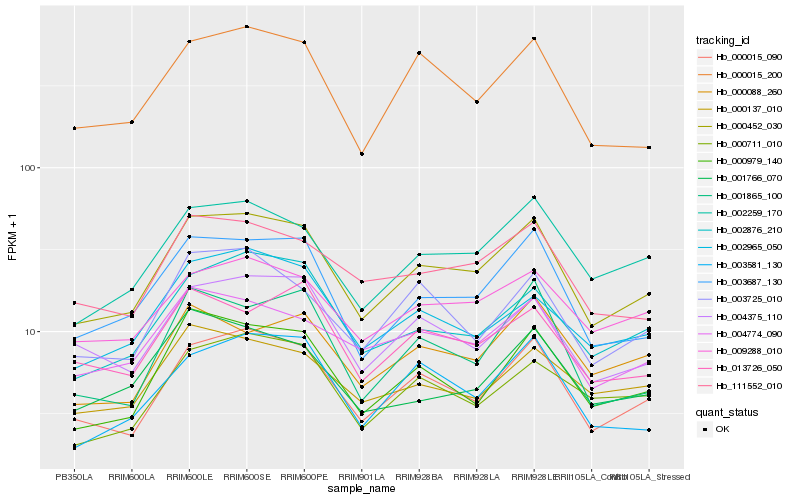
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000452_030 |
0.0 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 2 |
Hb_000015_090 |
0.057788163 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 3 |
Hb_003687_130 |
0.0635697103 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 4 |
Hb_002259_170 |
0.0741250668 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000979_140 |
0.079144925 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 6 |
Hb_000015_200 |
0.0828849223 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 7 |
Hb_111552_010 |
0.0847905729 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 8 |
Hb_004375_110 |
0.0851199161 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 9 |
Hb_002876_210 |
0.0861875129 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 10 |
Hb_009288_010 |
0.0879988755 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_004774_090 |
0.0882132484 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 12 |
Hb_002965_050 |
0.0936792978 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 13 |
Hb_003581_130 |
0.0941367228 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_003725_010 |
0.0942904892 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 15 |
Hb_000137_010 |
0.0962532615 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 16 |
Hb_001865_100 |
0.0970851021 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 17 |
Hb_000711_010 |
0.0979719549 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 18 |
Hb_001766_070 |
0.0984142207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_013726_050 |
0.0999720179 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 20 |
Hb_000088_260 |
0.1012274122 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |