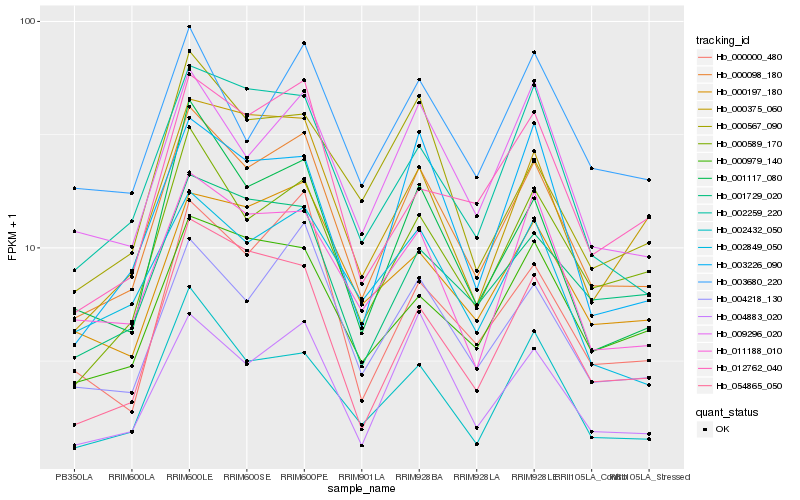
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004218_130 |
0.0808829613 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 3 |
Hb_000589_170 |
0.084584844 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_001117_080 |
0.0919278608 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009296_020 |
0.0984518727 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_002259_220 |
0.0994892031 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 7 |
Hb_011188_010 |
0.1009755066 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000979_140 |
0.1010498867 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 9 |
Hb_000197_180 |
0.1017392333 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 10 |
Hb_001729_020 |
0.1019940514 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000375_060 |
0.1050376205 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 12 |
Hb_054865_050 |
0.1069258075 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000000_480 |
0.1069260426 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002849_050 |
0.107315608 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 15 |
Hb_004883_020 |
0.1092364683 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 16 |
Hb_012762_040 |
0.1097481074 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 17 |
Hb_003680_220 |
0.1131903567 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 18 |
Hb_002432_050 |
0.1132500071 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 19 |
Hb_003226_090 |
0.1141498838 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 20 |
Hb_000567_090 |
0.1141780264 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |