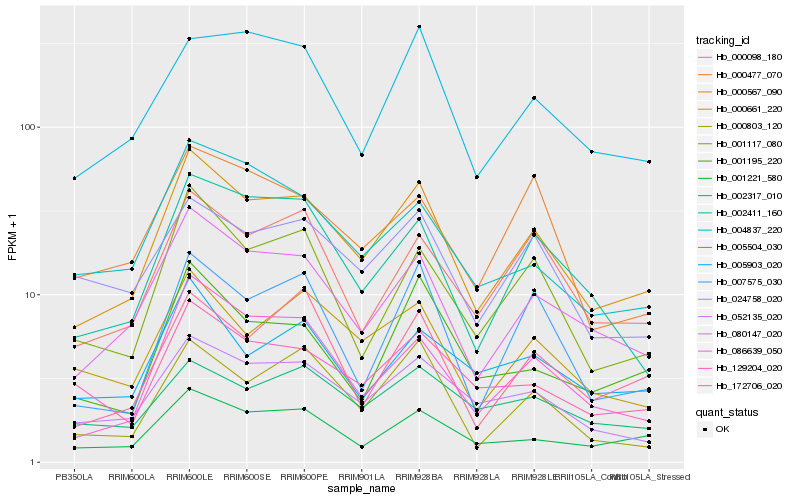
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_090 |
0.0 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_080147_020 |
0.0773515939 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 3 |
Hb_052135_020 |
0.1079492202 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 4 |
Hb_086639_050 |
0.108420684 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001221_580 |
0.1098748948 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 6 |
Hb_000098_180 |
0.1141780264 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001117_080 |
0.1157077209 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000661_220 |
0.117144315 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 9 |
Hb_002411_160 |
0.1185891539 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 10 |
Hb_129204_020 |
0.1254042012 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 11 |
Hb_005903_020 |
0.1283220344 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 12 |
Hb_005504_030 |
0.1290398539 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 13 |
Hb_024758_020 |
0.1313023424 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 14 |
Hb_000803_120 |
0.1314249217 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_007575_030 |
0.1341851022 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 16 |
Hb_001195_220 |
0.1342074822 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_004837_220 |
0.1344832474 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 18 |
Hb_002317_010 |
0.137491969 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_172706_020 |
0.1375556029 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 20 |
Hb_000477_070 |
0.1376095689 |
- |
- |
histone h2a, putative [Ricinus communis] |