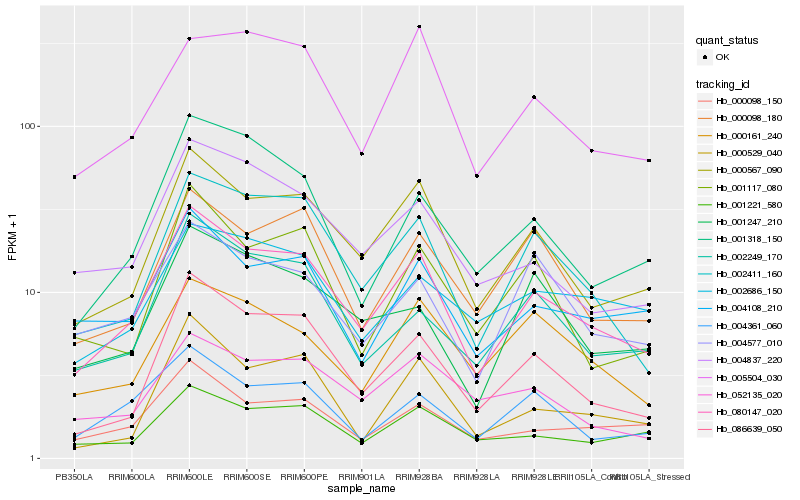
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080147_020 |
0.0 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 2 |
Hb_000567_090 |
0.0773515939 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_002411_160 |
0.0990917463 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 4 |
Hb_086639_050 |
0.1021346114 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000098_150 |
0.1155119367 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 6 |
Hb_001221_580 |
0.1192933776 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 7 |
Hb_004108_210 |
0.1201610599 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 8 |
Hb_004837_220 |
0.122526418 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 9 |
Hb_002249_170 |
0.1227791728 |
- |
- |
copine, putative [Ricinus communis] |
| 10 |
Hb_000098_180 |
0.1240775604 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004361_060 |
0.1242205675 |
- |
- |
PREDICTED: uncharacterized protein LOC105636705 [Jatropha curcas] |
| 12 |
Hb_005504_030 |
0.1292632054 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 13 |
Hb_052135_020 |
0.1300472059 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 14 |
Hb_001318_150 |
0.1307036333 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 15 |
Hb_004577_010 |
0.1321413965 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 16 |
Hb_001247_210 |
0.1329015347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001117_080 |
0.133268063 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000161_240 |
0.1353394108 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002686_150 |
0.1364027634 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_000529_040 |
0.137908756 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |