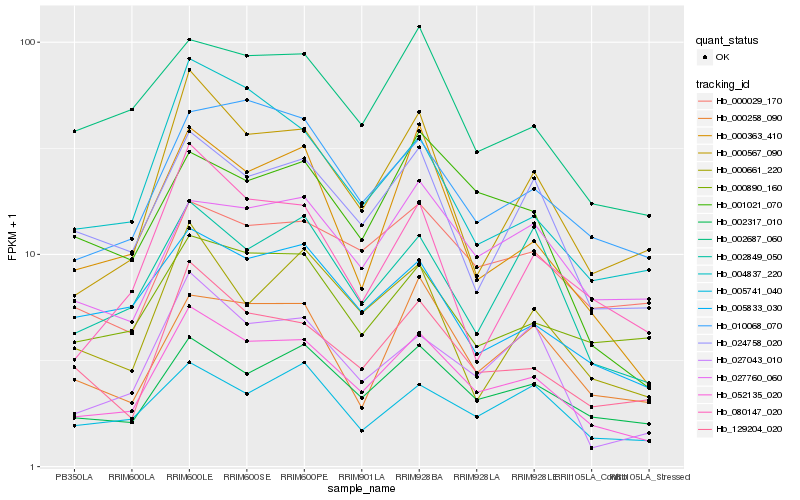
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052135_020 |
0.0 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 2 |
Hb_002317_010 |
0.094591899 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_129204_020 |
0.1006657995 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 4 |
Hb_000567_090 |
0.1079492202 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_010068_070 |
0.1079648813 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 6 |
Hb_024758_020 |
0.10982991 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 7 |
Hb_005833_030 |
0.1142688838 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 8 |
Hb_027043_010 |
0.1143435066 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase GDPD4 [Jatropha curcas] |
| 9 |
Hb_000363_410 |
0.1148157186 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 10 |
Hb_002687_060 |
0.1151960932 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 11 |
Hb_002849_050 |
0.1167409672 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 12 |
Hb_005741_040 |
0.1231348206 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 13 |
Hb_000890_160 |
0.1232045956 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 14 |
Hb_004837_220 |
0.1255255569 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 15 |
Hb_000258_090 |
0.1265474413 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000029_170 |
0.1274640072 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 17 |
Hb_001021_070 |
0.1274880855 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000661_220 |
0.128987329 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 19 |
Hb_027760_060 |
0.1292226556 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 20 |
Hb_080147_020 |
0.1300472059 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |