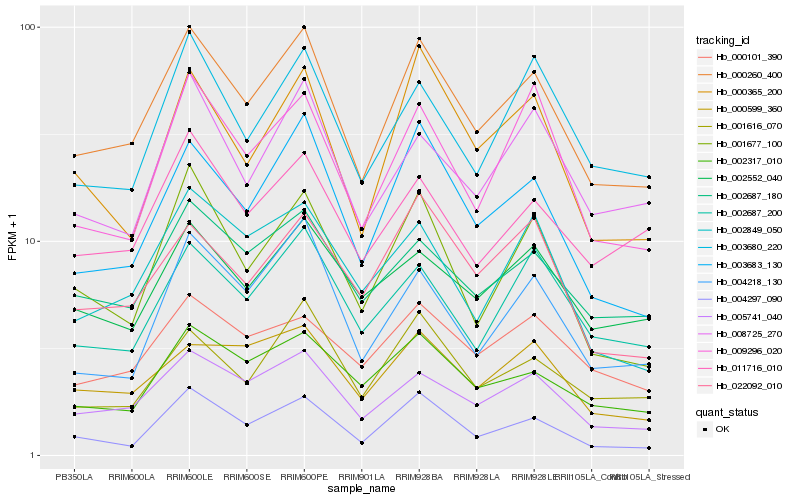
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_400 |
0.0 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 2 |
Hb_005741_040 |
0.0629127784 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 3 |
Hb_003683_130 |
0.0776591368 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002687_180 |
0.0891792153 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 5 |
Hb_002317_010 |
0.0916696502 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_001616_070 |
0.0945238184 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 7 |
Hb_002687_200 |
0.0994638188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000365_200 |
0.0999619649 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 9 |
Hb_000599_360 |
0.1008464994 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 10 |
Hb_009296_020 |
0.1015252256 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_003680_220 |
0.1015743485 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 12 |
Hb_022092_010 |
0.1016638033 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 13 |
Hb_002552_040 |
0.1020200591 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 14 |
Hb_001677_100 |
0.1020621502 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 15 |
Hb_000101_390 |
0.1030082591 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 16 |
Hb_004297_090 |
0.103361658 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 17 |
Hb_008725_270 |
0.1039105562 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 18 |
Hb_011716_010 |
0.1041591516 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 19 |
Hb_004218_130 |
0.1054081431 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 20 |
Hb_002849_050 |
0.1060944725 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |