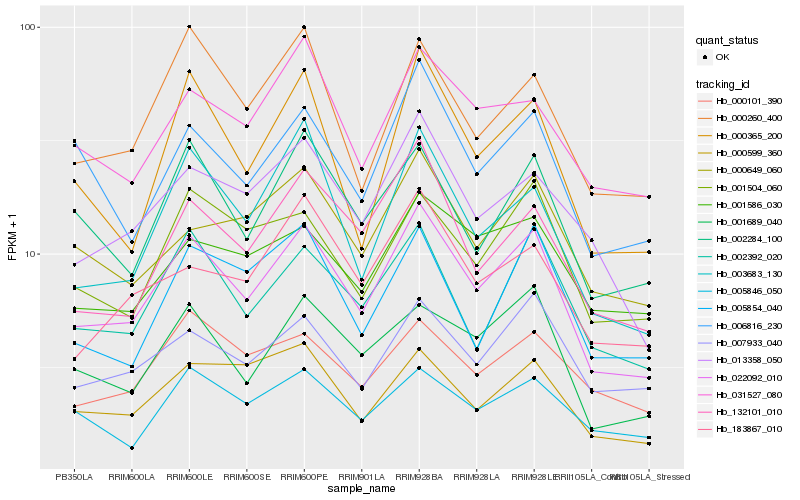
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022092_010 |
0.0 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 2 |
Hb_001689_040 |
0.0914005984 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_002392_020 |
0.0931654615 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 4 |
Hb_013358_050 |
0.096357759 |
- |
- |
NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_132101_010 |
0.0972240686 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 6 |
Hb_002284_100 |
0.099498337 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 7 |
Hb_000365_200 |
0.100245041 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 8 |
Hb_007933_040 |
0.1004679269 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003683_130 |
0.1006811254 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000260_400 |
0.1016638033 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 11 |
Hb_001504_060 |
0.1026505912 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 12 |
Hb_000599_360 |
0.1040754577 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 13 |
Hb_031527_080 |
0.1049087285 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 14 |
Hb_006816_230 |
0.1050097104 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 15 |
Hb_000649_060 |
0.1055718826 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 16 |
Hb_005846_050 |
0.1099017897 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 17 |
Hb_183867_010 |
0.1101378222 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_005854_040 |
0.1112244362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000101_390 |
0.1113984878 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 20 |
Hb_001586_030 |
0.1115647352 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |