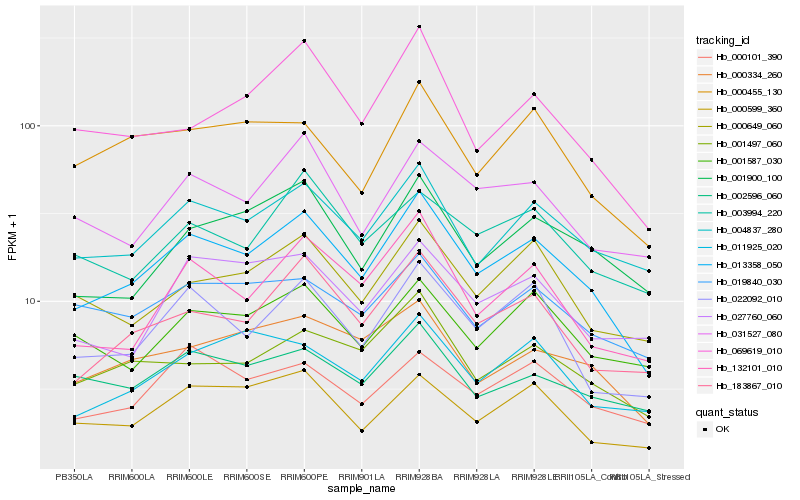
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013358_050 |
0.0 |
- |
- |
NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_004837_280 |
0.0929581399 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 3 |
Hb_183867_010 |
0.0942310923 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 4 |
Hb_022092_010 |
0.096357759 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 5 |
Hb_132101_010 |
0.0971713155 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 6 |
Hb_001497_060 |
0.1045606363 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 7 |
Hb_000334_260 |
0.1047471091 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 8 |
Hb_001900_100 |
0.1075065964 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000649_060 |
0.1090462929 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 10 |
Hb_000599_360 |
0.1093413939 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 11 |
Hb_031527_080 |
0.111376397 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 12 |
Hb_069619_010 |
0.1128500659 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 13 |
Hb_000101_390 |
0.1140676906 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 14 |
Hb_000455_130 |
0.1152655026 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002596_060 |
0.1170123136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_027760_060 |
0.1183262981 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 17 |
Hb_001587_030 |
0.1189215868 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 18 |
Hb_011925_020 |
0.1191019168 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 19 |
Hb_003994_220 |
0.1194740283 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 20 |
Hb_019840_030 |
0.1196059739 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |