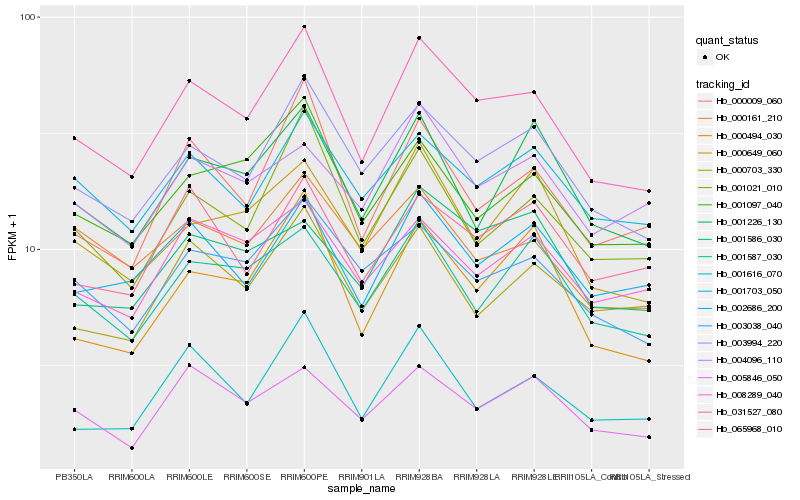
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031527_080 |
0.0 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 2 |
Hb_003994_220 |
0.0641197108 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 3 |
Hb_003038_040 |
0.0833356945 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 4 |
Hb_001021_010 |
0.0855299139 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 5 |
Hb_000009_060 |
0.0857363701 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 6 |
Hb_001097_040 |
0.0902549422 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 7 |
Hb_002686_200 |
0.0910065375 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 8 |
Hb_001703_050 |
0.0919623772 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 9 |
Hb_000494_030 |
0.0940044317 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 10 |
Hb_005846_050 |
0.0964029215 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 11 |
Hb_001587_030 |
0.096450196 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 12 |
Hb_000703_330 |
0.0974839838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000649_060 |
0.0976375989 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 14 |
Hb_000161_210 |
0.0976722578 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 15 |
Hb_008289_040 |
0.0984599998 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 16 |
Hb_065968_010 |
0.1008338055 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001226_130 |
0.1018543195 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 18 |
Hb_001586_030 |
0.1020017905 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 19 |
Hb_001616_070 |
0.1043379285 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 20 |
Hb_004096_110 |
0.1048466657 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |