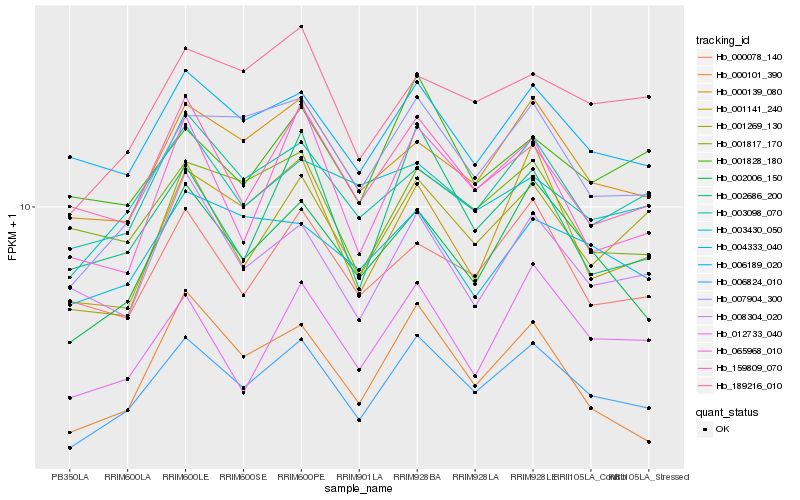
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006824_010 |
0.0 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 2 |
Hb_002006_150 |
0.0586914749 |
- |
- |
copine, putative [Ricinus communis] |
| 3 |
Hb_007904_300 |
0.0626030879 |
- |
- |
copine, putative [Ricinus communis] |
| 4 |
Hb_000139_080 |
0.0685884912 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006189_020 |
0.068964019 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 6 |
Hb_003098_070 |
0.0711940126 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 7 |
Hb_001269_130 |
0.0749033702 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 8 |
Hb_189216_010 |
0.0749924699 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 9 |
Hb_159809_070 |
0.0751035136 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 10 |
Hb_000078_140 |
0.0761370662 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 11 |
Hb_002686_200 |
0.0803780016 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 12 |
Hb_001817_170 |
0.0807961269 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 13 |
Hb_065968_010 |
0.0818120359 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 14 |
Hb_012733_040 |
0.0819101777 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 15 |
Hb_004333_040 |
0.0822097075 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 16 |
Hb_008304_020 |
0.0828613074 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 17 |
Hb_000101_390 |
0.0828645674 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 18 |
Hb_001141_240 |
0.0829699456 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001828_180 |
0.0844698195 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 20 |
Hb_003430_050 |
0.084603358 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |