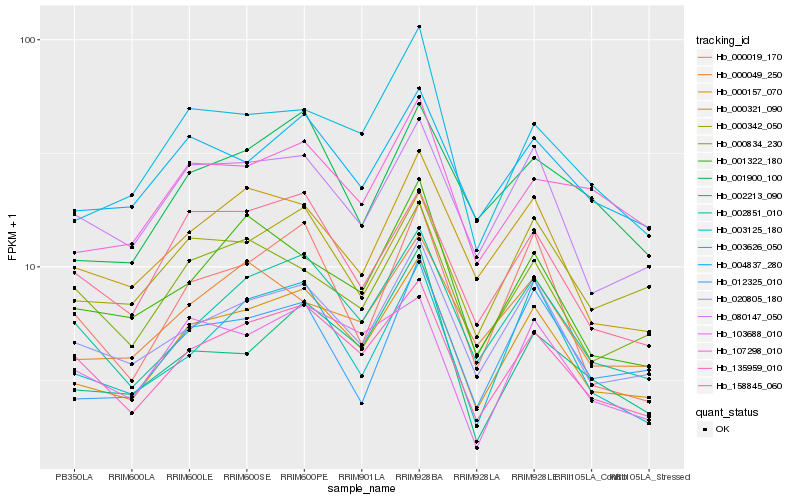
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000157_070 |
0.0 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 2 |
Hb_135959_010 |
0.0702356906 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 3 |
Hb_020805_180 |
0.0734811128 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 4 |
Hb_002851_010 |
0.0787564747 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 5 |
Hb_080147_050 |
0.0853091348 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 6 |
Hb_000342_050 |
0.0879737129 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 7 |
Hb_003626_050 |
0.0896343539 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 8 |
Hb_000321_090 |
0.0916075813 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 9 |
Hb_004837_280 |
0.0917233472 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 10 |
Hb_158845_060 |
0.0933654856 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003125_180 |
0.0951908556 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 12 |
Hb_012325_010 |
0.0994190815 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 13 |
Hb_001900_100 |
0.0995320578 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002213_090 |
0.1006626317 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 15 |
Hb_107298_010 |
0.1019615652 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 16 |
Hb_000019_170 |
0.102366028 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 17 |
Hb_001322_180 |
0.1026884495 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 18 |
Hb_103688_010 |
0.1028090968 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 19 |
Hb_000834_230 |
0.1034198858 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 20 |
Hb_000049_250 |
0.1034324021 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |